-
Tripartite motif (TRIM) family proteins, also named RBCC proteins, are characterized by three highly conserved domains, RING (really interesting new gene) finger, one or two B-box domains, and a coiled-coil region (McNab F W, et al., 2011). A number of TRIM family proteins exhibit E3 ubiquitin ligase activity, which is attributed to their RING domain. TRIM family proteins are involved in many cellular processes, including apoptosis, proliferation, oncogenesis, differentiation, intracellular trafficking, and innate immune responses (Ozato K, et al., 2008). In addition, many TRIM proteins have been reported to be able to regulate innate immunity through management of the nuclear factor-kappa B (NF-κB) pathway (Gong J, et al., 2011; Poole E, et al., 2009; Tian Y, et al., 2007; Zha J, et al., 2006; Zurek B, et al., 2012). TRIM30α can target transforming growth factor-β activated kinase (TAK) 1 binding protein 2 (TAB 2) and TAB 3 for degradation, and can negatively regulate the long terminal repeat (LTR)-mediated signal pathway (Shi M, et al., 2008). In glioblastoma, TRIM19, also named promyelocytic leukemia protein (PML), can inhibit the activation of the NF-κB pathway to activate caspase-8, then induce apoptosis (Kuwayama K, et al., 2009).
NF-κB is an important transcription factor that participates in cellular response to apoptosis, cellular proliferation, inflammation, and immune response. IκBα, one of the prototypical inhibitors of κB (IκB) proteins, plays an important role in mediating the canonical NF-κB pathway activation (Hayden M S, et al., 2008). Canonical NF-κB pathway activation requires phosphorylation of the IκB kinase (IKK) complex and the subsequent disassociation of IκBα from NF-κB. Moreover, IKK complex activation is regulated by tumor necrosis factor (TNF) receptorassociated factor 6 (TRAF6) and its downstream TRAF6-regulated IKK activator (TRIKA)1 and TRIKA2 protein complex (Wang C, et al., 2001). TRAF6, which has been characterized as an E3 ubiquitin ligase, can catalyze the lysine-63 (K63) polyubiquitination reaction that triggers IKK activation (Cao Z, et al., 1996). Furthermore, TRAF6 overexpression can activate the NF-κB pathway. This TRAF6-stimulated activation requires TRIKA1, a dimeric ubiquitin-conjugating enzyme complex composed of Ubc13 and Uev1A (Wang C, et al., 2001), and TRIKA2, another kinase complex consisting of TAK1, TAB 1, and TAB 2 (Besse A, et al., 2007). Activation of TAK1 kinase is induced by polyubiquitinated TRAF6. TAB 2, as a receptor, can preferentially bind to K63-linked polyubiquitin chains through a highly conserved zinc finger (ZnF) domain, which can facilitate TRAF6 self-ubiquitination and promote its assembly with IKK (Kanayama A, et al., 2004; Kishida S, et al., 2005).
TRIM22, a member of the TRIM/RBCC family, was first identified as an interferon-inducible gene in Daudi cells (Tissot C, et al., 1995). Studies have shown that TRIM22 is implicated in innate immunity (Barr S D, et al., 2008; Di Pietro A, et al., 2013; Gao B, et al., 2009), and TRIM22 has been identified as a p53 target gene involved in hematopoietic differentiation and T-lymphocyte activation (Obad S, et al., 2007; Obad S, et al., 2007). Recently, it was reported that TRIM22 overexpression activated the NF-κB pathway in unstimulated macrophage cell lines, and that this activation could be blocked by the IκBα inhibitorpyrollidine dithiocarbamate (PDTC) (Yu S, et al., 2011). However, the precise mechanisms of this activation are still unclear.
In the current study, we attempted to clarify this mechanism. We found that overexpressed TRIM22 negatively regulated the TRAF6-stimulated NF-κB pathway in the human embryonic kidney cell line HEK293T. TRIM22 inhibited TRAF6 self-ubiquitination, and this could be partially rescued by a TRIM22 RING domain deletion mutant. Additional experiments showed that TRIM22 was able to interact with TAB 2 and promote TAB 2 degradation, which might be responsible for inhibiting TRAF6 selfubiquitination.
HTML
-
The human TRIM22 gene and its deletion mutants were amplified by PCR from pCMV-SPORT6-TRIM22 (American Type Culture Collection (ATCC), Manassas, VA, USA) as described previously (Sivaramakrishnan G, et al., 2009). These sequences were cloned separately into the mammalian expression vector pcDNA3.1 (+) (Invitrogen Corp., Carlsbad, CA, USA) with a hemagglutinin (HA) epitope added to the N-terminal region of pLPCX (Clontech, Palo Alto, CA, USA) and a streptavidin tag (Strep-tag) added at its C-terminal. The resulting constructs were designated as TRIM22-WT (wild-type, with full-length TRIM22), TRIM22-ΔR (RING-domain deletion mutant) and TRIM22-ΔS (SPRY-domain deletion mutant). The ubiquitin gene was amplified with the HA epitope added to its N-terminus from the cDNA of HeLa cells as described previously (Huang F, et al., 2013). We also used a NF-κB-luciferase reporter plasmid (pNF-κB-luc; Agilent Biotechnology, Santa Clara, CA, USA) and an RL-TK-luciferase (pRL-TK-luc) reporter plasmid (Promega Corp., Madison, WI, USA).
-
The antibodies used were anti-HA and anti-Flag antibodies (Sigma-Aldrich, St. Louis, MO, USA), and anti-Strep and goat anti-mouse IgG (horseradish peroxidase (HRP)-conjugated) antibodies (Cell Signaling Technology, Danvers, MA, USA).
-
HEK293T cells were cultured with Dulbecco's modified Eagle's medium (DMEM) (Gibco, Auckland, NZ) supplemented with 10% fetal calf serum (FBS) (Gibco). Transfection of the cells was maintained with transfection reagent (LipofectamineTM 2000 CD Reagent; Invitrogen) following the manufacturer's protocol.
-
HEK293T cells (1×105) were maintained in 24-well dishes and transfected the following day with the indicated plasmids, while pPL-TK-luc with pNF-κB-luc were co-transfected into the cells to normalize the transfection efficiency. The cells were harvested 24 h later, and NF-κB activity was analyzed using a commercial assay (Dual Luciferase® Reporter Assay System (Promega)) following the manufacturer's instructions.
-
We performed a two-step immunoprecipitation assay to detect the ubiquitination level of TRAF6. After cells transfected with the Flag-tagged TRAF6 and HA-tagged ubiquitin plasmids were co-transfected with or without the indicated TRIM22 recombinant plasmid for 36 h, the cells were lysed with lysis buffer (20 mmol/L Tris, pH 7.5, 150 mmol/L NaCl, 1% TritonX-100,1 mmol/L EDTA, 10 mg/mL aprotinin, 10 mg/mL leupeptin, and 1 mmol/L phenyl-methyl-sulfonyl fluoride). Firstly, the cell lysates were immunoprecipitated with resin (Protein G-Sepharose; GE Healthcare, Piscataway, NJ, USA) and indicated antibody, then the immunoprecipitates were treated as described previously (Zhong B, et al., 2010). Secondly, the immunoprecipitates were re-immunoprecipitated with magnetic beads (Anti-FLAG M2 Gel Beads; Sigma-Aldrich), following the manufacturer' instructions. The immunoprecipitates were then analyzed by western blotting with the indicated antibodies (Gong J, et al., 2011; Huang F, et al., 2013).
-
For the co-immunoprecipitation (Co-IP) assay, HEK293T cells (2×106) were transfected in six-well dishes with indicated plasmids. At 24 h post-transfection, cells were lysed with lysis buffer on ice for 20 min. The cellular debris was cleared by centrifugation at 15,000×g for 10 min at 4℃. The lysates were then incubated with anti-FLAG M2/anti-HA affinity gel beads (Sigma-Aldrich) for 4 h at 4℃. Finally, the samples were treated as described above for the ubiquitination assay.
Plasmid constructs and deletions
Antibodies
Cell culture and transfection
Dual luciferase assay
In vivo ubiquitination assay
Co-immunoprecipitation assay
-
It has been reported that TRIM22 activates the NF-κB pathway in macrophage cell lines, and this is closely linked with the IκBα dissociation from NF-κB (Yu S, et al., 2011). The results of the current study confirmed that TRIM22 was able to activate the NF-κB pathway in a dose-dependent manner in HEK293T cells (Fig. 1A). However, when TRIM22 and TRAF6 were co-transfected, TRIM22 downregulated, in a dose-dependent manner, the NF-κB pathway activation that was stimulated by TRAF6 (Fig. 1B). These results suggest that TRIM22 can inhibit TRAF6-induced NF-κB pathway activation.
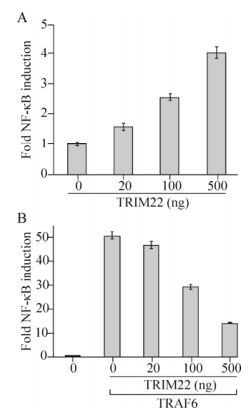
Figure 1. TRIM22 regulates the NF-κB pathway. HEK293T (1×105) cells were transfected with 0.05 μg of pNF-κB-luc and pRL-TK-luc and increasing amounts of HA-TRIM22 (A), or 0.1 μg of Flag-TRAF6 and increasing amounts of HA-TRIM22 (B). After 24 h, cells were lysed and used in a dual luciferase assay. Results were standardized to 1 in cells transfected with vector. For luciferase assays, data (means and SEM) presented are representative of at least three independent experiments.
-
To investigate the mechanism by which TRIM22 downregulates the TRAF6-induced NF-κB pathway activation, we constructed TRIM22 with a RING domain deletion (ΔR) and a SPRY domain deletion (ΔS) (Fig. 2A). The dual luciferase assay was performed in cells transfected with or without TRAF6 and TRIM22 or its mutant ΔR, and the results indicated that the inhibition caused by wild-type TRIM22 could be partially rescued by the mutant ΔR (P < 0.05) (Fig. 2B). It is known that TRAF6 self-ubiquitination is crucial for NF-κB pathway activation; however, to determine whether TRIM22 affects the level of TRAF6 self-ubiquitination, we assessed whether ubiquitinated TRAF6 could be detected in the cells transfected with TRAF6 and TRIM22 or its mutant ΔR. As shown in Fig. 2C, TRIM22 (lane 3) was able to decrease the level of self-ubiquitinated TRAF6, and the TRIM22 mutant ΔR (lane 4) was able to partially rescue this decrease. Our results suggest that ΔR can partially rescue the inhibition of TRAF6-induced NF-κB pathway activation by reversing of the reduction in TRAF6 self-ubiquitination caused by wild-type TRIM22.
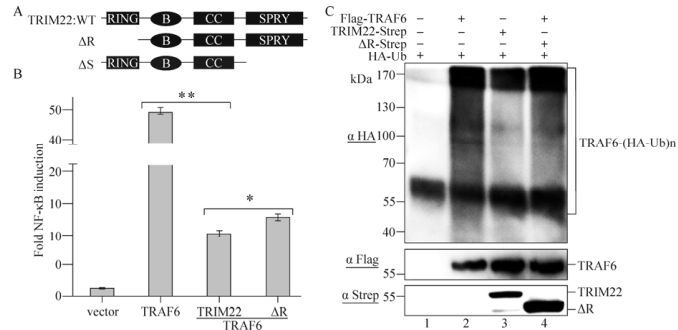
Figure 2. TRIM22 affects the self-ubiquitination of TRAF6. A: Schematic representation of wild-type TRIM22 (WT), the RING domain deletion mutant (ΔR), and the SPRY domain deletion mutant (ΔS). B: HEK293T cells (1×105) were transfected with 0.05 μg of pNF-κB-luc and pRL-TK-luc, 0.1 μg of Flag-TRAF6, and vector/TRIM22/ΔR. After 24 h, cells were lysed and used in a dual luciferase assay. Results are standardized to 1 in cells transfected with a vector. For luciferase assays, the data (means and SEM) are representative of three independent experiments. *P < 0.05, **P < 0.01. C: HEK293T cells were transfected with HA-Ub and/or Flag-TRAF6 (lanes 2 and 1) in the presence of TRIM22/ΔR (lanes 3 and 4) or not. After 36 h, cells were lysed for ubiquitination assay using the two-step immunoprecipitated strategy, then ubiquitinated TRAF6 was detected by the anti-HA antibody (upper panel) and Flag-tagged TRAF6 and Strep tagged TRIM22/ΔR were separately detected by anti-Flag and anti-Strep antibody in immunoprecipitates (middle panel) and cell lysates (input, lower panel).
-
In a previous report (Kishida S, et al., 2005), TAB 2 was identified as an adaptor that links TAK1 to an upstream signaling intermediate, TRAF6, which can facilitate TRAF6 self-ubiquitination. To determine whether TRIM22 can inhibit self-ubiquitination of TRAF6 by regulating TAB 2 levels, TAB 1 and TAB 2 were co-transfected with TRIM22 into HEK293T cells. As shown in Fig. 3A, compared with TAB 1 (lanes 3 and 4), the level of TAB 2 was significantly decreased by TRIM22 (lanes 5 and 6). Furthermore, this decrease occurred in a dose-dependent manner (Fig. 3B). Additionally, as shown in Fig. 3C, the TRIM22 mutant ΔR was able to partially rescue the decrease in TAB 2 levels caused by wild-type TRIM22. To determine the detailed mechanism of TAB 2 degradation by TRIM22, three different types of inhibitors (MG132, NH4Cl, and chloroquine (CHQ)) were individually added to the cell-culture medium. As shown in Fig. 3D, compared with the DMSO control (lanes 1 and 2), NH4Cl but not MG132 (lanes 3 and 4) or CHQ (lanes 7 and 8) prevented TAB 2 degradation by TRIM22 (lanes 5 and 6). These results demonstrate that TRIM22 can induce TAB 2 degradation, and the TRIM22 mutant ΔR can partially rescue this degradation.
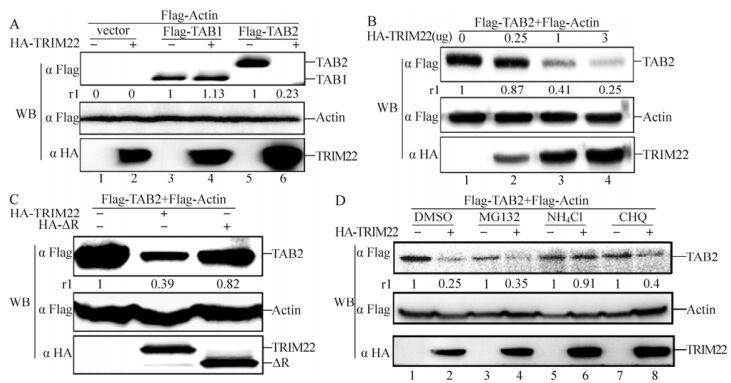
Figure 3. TRIM22 targets TAB 2 for degradation. HEK293T cells (2×106) were transfected with a vector (lanes 1 and 2), Flag-TAB 1 (lanes 3 and 4), Flag-TAB 2 (lanes 5 and 6), or Flag-actin in the presence or absence of TRIM22. HEK293T cells (2×106) were transfected with Flag-TAB 2, Flag-actin, or increasing amounts of HA-TRIM22. After 30 h, cells were treated as described above. C: HEK293T cells (2×106) were transfected with Flag-TAB 2 and Flag-actin in the presence of HA-TRIM22 or HA-ΔR (lanes 2 and 3) or in their absence (lane 1). After 30 h, cells were harvested and lysed, and the cell lysates were subjected to western blotting and probed with indicated antibodies. D: HEK293T cells (2×106) were transfected with Flag-TAB 2 and/or HA-TRIM22. After 24 h, the cells were treated with or without DMSO (lanes 1 and 2), 20 mmol/L MG132 (lanes 3 and 4), 5 mmol/L NH4Cl (lanes 5 and 6), or 10 mmol/L chloroquine (CHQ) (lanes 7 and 8) for 6 h. Next, western blotting was performed with the indicated antibodies. All data are representative of at least three independent experiments; the relative levels (rl) of the indicated proteins to actin expression level in these experiments are presented at the bottom of the indicated panels.
-
Several other members of the TRIM family, such as TRIM30α, have been shown to interact with TAB 2 (Shi M, et al., 2008). To determine whether the TRIM22-driven TAB 2 degradation is linked to a similar interaction, we co-transfected TAB 1 or TAB 2 with TRIM22 for the Co-IP assay. As shown in Fig. 4A, TAB 2 was immunoprecipitated by TRIM22 (lane 2), whereas TAB 1 was not (lane 1). Furthermore, to confirm which region of TRIM22 is necessary for TAB 2 interaction, the TRIM22 or TRIM22 mutants (ΔR and ΔS) were co-transfected with or without TAB 2 for the Co-IP assay. As shown in Fig. 4B, TRIM22 (lane 3) and its two mutants (ΔR and ΔS) (lanes 5 and 7) all interacted with TAB 2. These results show that TRIM22 can interact with TAB 2, and this interaction is independent of the RING or SPRY domains but might be dependent on its BC (B-box domain and coiled-coil region) domain.
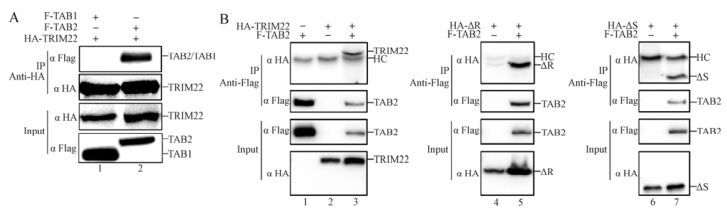
Figure 4. TRIM22 interacts with TAB 2. HEK293T cells (2×106) were transfected with HA-TRIM22 and Flag-TAB 1 (lane 1) or Flag-TAB 2 (lane 2) (A). HEK293T cells (2×106) were transfected with/without Flag-TAB 2 (lane 1) and HA-TRIM22 (lanes 2 and 3)/ HA-ΔR (lanes 4 and 5)/HA-ΔS (lanes 6 and 7) (B). After 30 h, the cells were harvested and lysed, then the cell lysates were immunoprecipitated with anti-HA (A) or anti-Flag (B). The HA-tagged TRIM22/mutants and Flag-tagged TAB 2 in the immunoprecipitates (upper panel) and cell lysates (input) (lower panel) were detected with the indicated antibodies.
TRIM22 downregulates the TRAF6-stimulated NF-κB pathway
TRIM22 inhibits self-ubiquitination of TRAF6 through its RING domain
TRIM22 targets TAB 2 for degradation, depending on its RING domain
TRIM22 and TAB 2 interaction is not dependent on the RING or SPRY domains
-
Many members of the TRIM family have been shown to play important roles in innate immunity (Ozato K, et al., 2008), and certain TRIMs can modulate various stages of the NF-κB pathway (Poole E, et al., 2009; Tian Y, et al., 2007; Zha J, et al., 2006; Zurek B, et al., 2012). TRIM22 has been reported to activate the NF-κB pathway in macrophage cell lines (Yu S, et al., 2011), but it is still unknown which NF-κB pathway stage is affected by TRIM22. In this study, we confirmed that overexpressed TRIM22 was able to activate the NF-κB pathway in HEK293T cell lines and was also able to inhibit the TRAF6-induced NF-κB pathway activation. Having identified TRIM22 as a negative modulator of the TRAF6-activated NF-κB pathway, we also showed that it induced TAB 2 degradation and thereby inhibited TRAF6 self-ubiquitination.
TRAF6, an E3 ligase that serves as a scaffold to connect the upstream and downstream signaling proteins, is an important signaling adapter protein for NF-κB activation (Cao Z, et al., 1996). Moreover, TRAF6 self-ubiquitination, with K63-linked chains, is pivotal to NF-κB activation (Hayden M S, et al., 2008). According to a previous report (Zhang X, et al., 2013), UBE2O can bind to TRAF6 to inhibit its K63 polyubiquitination, and prevent NF-κB activation by interleukin (IL)-1β and lipopolysaccharide. In the current study, we also found that TRIM22 overexpression could inhibit the TRAF6-stimulated NF-κB pathway, and that this inhibition could be partially rescued by a TRIM22 RING domain deletion mutant (Fig. 1). Consistent with this phenomenon, when plasmids containing TRIM22 or TRAF6 were co-transfected into HEK293T cells, TRIM22 also decreased the self-ubiquitination level of TRAF6, and this was dependent on the RING domain of TRIM22 (Fig. 2). These results indicate that TRIM22 inhibits NF-κB pathway activation by decreasing TRAF6 self-ubiquitination. Additionally the TRIM22 ΔR mutant could rescue self-ubiquitination only to a certain extent; it was unable to return the levels to those without TRIM22. These findings imply that, TRIM22 might affect other components that facilitate TRAF6 self-ubiquitination.
The TRIKA2 protein complex, which consists of TAK1, TAB 1, and TAB 2, is the primary signaling protein downstream of TRAF6 in the IL-1β or TNF-α-stimulated NF-κB pathway (Besse A, et al., 2007). TAB 2 has been identified as a promoter of TRAF6 self-ubiquitination, which could, in turn, increase NF-κB activation (Kishida S, et al., 2005). In our study, we found that, compared with TAB 1, TRIM22 was able to interact with and degrade TAB 2 (Fig. 4A and Fig. 3A), similar to the TRAF6-stimulated NF-κB pathway inhibition by overexpressed TRIM22 described above. Additionally, the TRIM22 mutant ΔR was able to partially rescue the TAB 2 degradation caused by wild-type TRIM22 (Fig. 3C). Furthermore, the results of the Co-IP assay using two TRIM22 mutants (ΔR and ΔS) and wild-type TRIM22 indicated that the BC domain of TRIM22 might be pivotal to the interaction between TRIM22 and TAB 2.
In conclusion, we found that overexpression of TRIM22 can negatively regulate the TRAF6-stimulated NF-κB pathway in HEK293T cells, and that TAB 2 might be a key component in this regulation. TRIM30α, a negative regulator of the NF-κB pathway, is crucial for downregulation of the inflammatory response (Wimmer N, et al., 2012). Considering both the NF-κB pathway activation by TRIM22 and the TRAF6-stimulated NF-κB pathway inhibition by TRIM22, it is possible that TRIM22 is important in balancing the innate immune response.
As the inhibition of NF-κB pathway shown here is based on the overexpression of TRIM22 and TRAF6, it will be necessary in the future to confirm our results by performing knock-down experiments to verify whether endogenous TRIM22 can also downregulate the TRAF6-mediated NF-κB pathway. In addition, endogenous experiments need to be performed to confirm whether the interaction does exist between endogenous TRIM22 and TAB 2, and whether TAB 2 can be degraded by endogenous TRIM22. Finally, as many stimuli can activate the NF-κB pathway through TRAF6 and TAB 2, further experiments should focus on the identification of the exact pathway type involving TRIM22.
-
We thank Dr. Hong-Bing Shu (Wuhan University, Wuhan, China) for providing the plasmid of Flag-TAB 2 and Dr. Yan-Yi Wang (Wuhan Institute of Virology, CAS) for providing the plasmid of Flag-TRAF6. This work was supported by a grant from the Key Projects in the National Science & Technology Pillar Program during the twelfth Five-Year Plan Period of China (2012ZX10001006-002) and grants from the International Science & Technology Cooperation Program of China (2011DFA31030) and Deutsche Forschungsgemeinschaft (SFB/Transregio TRR60) and Key Laboratory on Emerging Infectious Diseases and Biosafety in Wuhan.
-
Conceived and designed the experiments: HQ, BL S and RG Y. Performed the experiments: HQ, FH and HX. Analyzed the data: HQ, FH and BL S. Contributed reagents/materials: HQ and HX. Wrote the paper: HQ, FH, BL S and RG Y.













 DownLoad:
DownLoad: