HTML
-
The HIV-1 infection in humans has now become a pandemic and in the absence of treatment it often leads to acquired immunodeficiency syndrome (AIDS). Although significant progress has been made to reduce the HIV-1 viral load in the blood of infected patients, challenges still exist in areas such as viral latency and viral resistance (Hosseinipour M C, 2013; Katlama C, 2013). To flush out or eradicate HIV-1 from the patients, scientists are examining the host factors that HIV-1 relies on for its replication (Shirakawa K, 2013; Walker B D, 2013). Some of these host factors have been identified in the past two decades by exploring the species differences of HIV-1 replication. For example, in humans, but not in the mouse, CD4 functions as the receptor for the entry of HIV-1 into the host cells. Other well-known host factors include CXCR4, CCR5, and Cyclin T1. However, mouse primary T cells expressing these human proteins failed to efficiently produce infectious HIV-1, indicating that additional host factors remain to be identified (Zhang J X, 2008).
Functional genetic screening using RNAi technology has been one of the most important tools in identifying host factors that regulate HIV replication. These studies (Brass A L, 2008, Zhou H, 2008) often used replication-competent viruses and required stringent personal protection; thus, adoption and validation under normal laboratory settings was limited. Moreover, many of these screening strategies failed to pinpoint the host factors supporting both the early and late stages of the HIV-1 life cycle. Unless huge efforts were invested in following-up on the likely hits, these screens could miss out a lot of important information. In the present report, we have established two RNAi library screening assays that: 1) utilize replication incompetent HIV-1 viruses and 2) identify host factors involved in the early stage and late stages of the HIV life cycle.
Our first screening assay (Fig. 1) was designed to identify host factors involved in the early stage of the HIV life cycle, till the Tat mediated trans-enhancement, which includes viral entry, reverse transcription, nuclear entry, integration, transcription, and trans-enhancement. The TZM-bl cells that were chosen for this assay are a HeLa cell line stably expressing hCD4, CXCR4, CCR5 to support the entry of HIV-1, as well as a luciferase reporter under the control of HIV-1 LTR, which is activated only after the virus has completed all the steps in the early stage of the HIV life cycle to express the Tat protein (Platt E J, 2009).
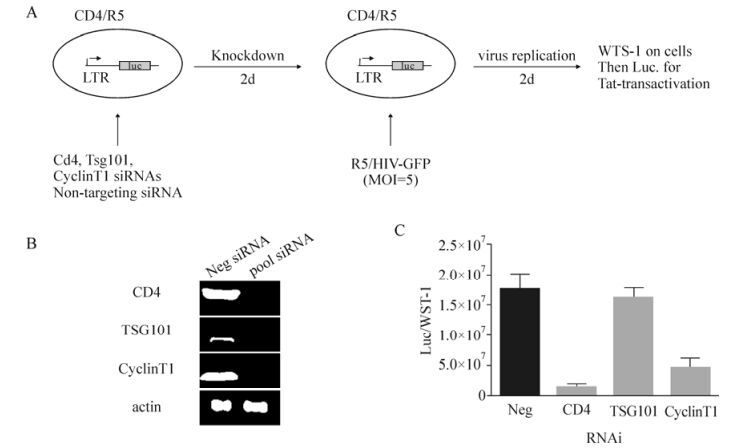
Figure 1. siRNA library screening strategy for the entry of HIV-1 for trans-enhancement. A) Schematic representation of the screening strategy for HIV entry for trans-enhancement; B) Knockdown efficiencies of siRNAs targeting CD4, TSG101, and Cyclin T1, with a non-targeting siRNA as the negative control; C) Effects of siRNAs. Error bars indicate the standard deviation from experiments conducted in triplicate. LTR, long terminal repeat; Luc, luciferase; Neg, negative control.
siRNAs against CD4 and Cyclin T1 were chosen as positive controls based on their recognized function during viral entry and transcription (Wei P, 1998), whereas siRNAs against Tsg101, which is involved in the late stage of the HIV life cycle (Garrus J E, 2001), served as a negative control. The window for the siRNA knockdown was optimized at two days post transfection to allow both, sufficient time for protein downregulation and to minimize cell death resulted from the knockdown of essential genes. For example, we found that the knockdown of Cyclin T1 for more than five days resulted in increased cell death.
In this assay, the TZM-bl cells transfected with the respective siRNAs were infected with a replication incompetent R5/HIV-GFP virus that lacked the Env gene and the virus was pseudotyped with a CCR5-tropic Env protein. Two days following the viral infection, the cells were subjected to a WST-1 viability assay, followed by a luciferase assay to assess the LTR promoter activity indicating the completion of the early life cycle of HIV-1. WST-1 was selected in this assay because it showed minimal effect on the luciferase signals and correlated well with cell numbers in the range of 0-10,000 cells.
As shown in Fig. 1, the knockdown of CD4 reduced the luciferase signals up to 10-fold, whereas the knockdown of Cyclin T1 reduced the signal up to 3-fold; the Tsg101 knockdown showed little effect as expected. This screening assay required a total of 5 days, starting with the siRNA transfection to the luciferase measurement.
Our second screening assay was designed to identify host factors associated with the late stage of the HIV-1 life cycle, including viral transcription, translation, Gag assembly, virus release to maturation. One of the major challenges in this assay was the selection of the virus to meet the issues of both safety and an accurate readout. An ideal virus would be replication incompetent for reasons of safety, could be regulated in its expression to minimize HIV toxicity in stable cell lines, and would be infectious to allow the use of virus infectivity as the readout.
One of the strategies for such a viral system was the separation of HIV-1 into Rev and Rev-deficient HIV-1. Regulation of Rev protein expression was expected to control the release of infectious viruses; while this was true in the 293T cells co-transfected with Rev and Rev-deficient HIV-1 (Sadaie M R, 1988), inconsistent results were observed in cell lines stably expressing the Rev protein.
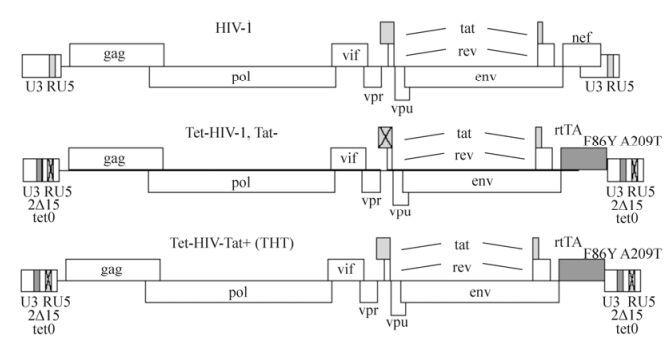
We identified that the best strategy was to control the HIV-1 LTR promoter activity with a Tet inducible element. In Tet-HIV (Das A T, 2005), tetO elements were introduced in LTRs and the Nef gene was replaced with a rtTA; thus the LTR promoters were doxycycline inducible. However, in this construct, the Tat gene was mutated rendering the construct unsuitable for the infectivity assay. To reintroduce the Tat gene into the construct, the sequence of the wild type Tat was excised from the LAI strain of HIV-1 using the restriction enzymes DraⅢ and NcoI, and cloned into the same sites in Tet-HIV; the resulted virus construct was named as Tet-HIV-Tat (THT) (Fig. 2). An infectious virus could only be generated in the presence of doxycycline. In addition to being replication incompetent, this construct has two other unique features: 1) Tat expression is independent of doxycycline, indicating that to analyze the viral infectivity with the LTR-Luc reporter in the TZM-bl cells, no addition of doxycycline was necessary. This made the infectivity assay safer because no secondary infectious virus was produced from the TZM-bl cells. 2) Although TAR was mutated in THT-HIV, Cyclin T1 was still necessary for virus production, which could be due to its role in protein translation (Das A T, 2007). This presented Cyclin T1 as a positive control in this screening strategy.
The GHOST cell line chosen for this assay was a human osteosarcoma cell line expressing a LTR-driven GFP reporter. The cells were initially infected with the THT virus pseudotyped with the VSVG envelope to establish the GHOST-THT stable cell line by selecting for the GFP positive population, which indicated the presence of the integrated THT virus. Importantly, no toxicity was observed in the stable cell lines established using the HIV construct because the expression was only after induction. The cells were transfected with various siRNAs and allowed 2 days for an efficient knockdown. Doxycycline was then added to induce the release of the infectious but replication incompetent THT virus (Fig. 3). One day later, the supernatant containing the THT virus was transferred to the TZM-bl cells to assay for viral infectivity, whereas the GHOST-THT cells were subjected to a viability assay as indicated above.
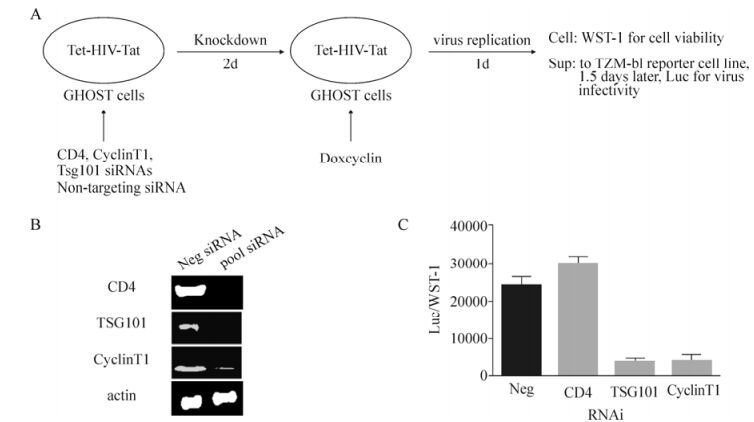
Figure 3. siRNA library screening strategy for virus release. A) Schematic representation of the screening strategy for virus release; B) Knockdown efficiencies of siRNAs targeting CD4, TSG101, and Cyclin T1, with non-targeting siRNAs as negative control; C) Effects of siRNAs. Error bars indicate the standard deviation from experiments conducted in triplicate.
As shown in Fig. 3, the knockdown of Cyclin T1 for two days inhibited the viral release by up to 5-fold. Tsg101 is critical for HIV-1 release and served as another positive control in our study. This screening assay took four days, beginning with the siRNA transfection to the luciferase assay and viability readout. We are currently validating multiple host factors reported in the literature, including lens epithelium-derived growth factor (LEDGF), HIV-1 Tat specific factor1 (HTATSF1), Cdk9, retrograde Golgi transport proteins (Rab6 and Vps53), karyopherin (TNPO3), and the Mediator complex (Med28).
In summary, two RNAi library screening assays have been successfully developed that utilize replication incompetent HIV-1 viruses to identify host factors that are independently involved in the early and the late stages of the HIV life cycle. Such assays would prove helpful in identifying important targets for drug development to eradicate HIV.











 DownLoad:
DownLoad: