HTML
-
Enterovirus 71(EV71), a member of the genus Enterovirus of the family Picornaviridae, is a single-str and ed, positive-sense RNA virus that usually causes mild h and foot-mouth disease(HFMD)in children, with symptoms such as fever, diarrhea, and herpangina(Liu et al., 2013). However, certain strains of EV71 infection can cause severe neurological complications, such as SDLY107(Sun et al., 2014). EV71 is classified into three distinct genotypes(A–C); the B and C genotypes are further divided into B1–B5 and C1–C5 genotypes, respectively, based on the VP1 sequence. Among these subgenotypes, C4 is the predominant circulating genotype in China(Huang et al., 2010). The genomic RNA of EV71, flanked by a 5′ untranslated region(UTR) and a 3′UTR, is approximately 7, 500 nucleotides in length and encodes a large polyprotein, which is autocatalytically cleaved into four structural proteins(VP1–VP4) and seven nonstructural proteins(2A–2C and 3A–3D). Similar to the genomes of other picornaviruses, the EV71 genome possesses a 5′UTR of approximately 745 nucleotides and can be divided into two secondary structures: the stem-loop Ⅰ and the internal ribosome entry site(IRES)elements(comprising stemloops Ⅱ–Ⅵ)(Deng et al., 2012; Thompson and Sarnow, 2003). The stem-loop Ⅰ element at the N-terminus of the 5′UTR plays an important role in viral RNA replication, and the IRES element can initiate genome translation through an IRES-dependent translation mechanism(Luo et al., 2014).
The significant morbidity and mortality rates associated with frequent outbreaks of EV71 worldwide have raised public health concerns(Zhang et al., 2010). In a large population survey conducted from 2008 to 2014, EV71 was found to infect 10, 717, 283 individuals, causing 3046 deaths(Liu et al., 2015). Despite improvements in virological and medical research, no specific drugs or vaccines are commercially available for the prevention or treatment of EV71 infection owing to our lack of adequate underst and ing of the pathogenic mechanism of this virus. Therefore, there is an urgent need to investigate the pathogenesis of EV71.
In a previous study, we demonstrated that the replication capacity differed between high and low virulence strains of EV71, indicating that differences in replication capacity may be associated with the pathogenic mechanisms of EV71(Sun et al., 2014). However, the region in the EV71 genome governing the differential replication capacities of these strains has not yet been explored. In this study, we constructed and characterized infectious cDNA clones of EV71: the rescued full-length SDLY107(named SDLY107RV) and rescued recombinant infectious cDNA clones(named SDLY107(1–5′UTR)RV). The role of the EV71 5′UTR in controlling the replication of the virus was further explored.
SDLY107(accession number: JX244186.1) and SDLY1(accession number: JX244182.1), which clustered into C4a of the C4 subgenotype, were isolated from children with HFMD collected from Linyi People's Hospital(Wen et al., 2013). To construct infectious cDNA clones of EV71, we amplified the open reading frame(ORF)segment from the cDNA template of SDLY107 by polymerase chain reaction(PCR)with primers ORF107F and ORF107R.(Sequences of all primers are in Supplemental Table S1.)The resulting DNA fragment, double digested with Xba Ⅰ and Hind Ⅲ, was ligated into the pcDNA3.1(+)plasmid. The 5′-terminal sequence of SDLY107, containing approximately 1.5 kb, was amplified by PCR with the primers 5′UTR107 F and VP4 107 R and inserted into the above construct following double digestion with Hind Ⅲ and Bsi WI. The 3′-terminal sequence of SDLY107, containing approximately 1.6 kb, was amplified by PCR with primers 3′UTR Bsp1407Ⅰ F and 3′UTR52T R and cloned into the above construct following double digestion with Bsp1407 Ⅰ and Xba Ⅰ to yield pcDNA3.1(+)SDLY107(Figure 1A). In addition, by replacing the 5′UTR of SDLY107 with that of SDLY1, the recombinant infectious cDNA clones pcDNA3.1(+)SDLY107(1–5′UTR)were constructed(Figure 1A). Sequence analysis revealed that pcDNA3.1(+)SDLY107 and pcDNA3.1(+)SDLY107(1–5′UTR)were constructed successfully and that there were no nucleotides differing from the previously reported EV71 strain SDLY107 or SDLY1(data not shown).
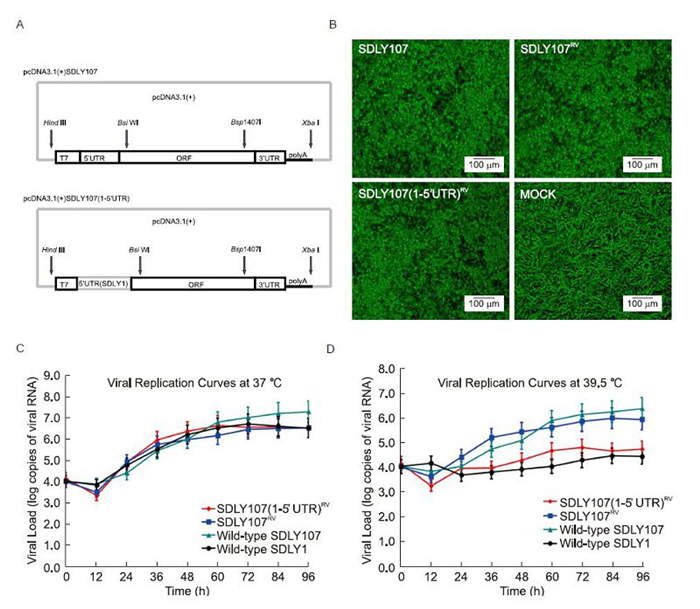
Figure 1. (A) Schematic diagram of pcDNA3.1(+)SDLY107 and pcDNA3.1(+)SDLY107(1–5′UTR). (B) The cytopathic effects of infection with wild-type SDLY107, SDLY107RV, or SDLY107(1–5'UTR)RV or mock infection in RD cells at 24 h post-transfection were assessed by microscopy (200× magnification). (C, D) The SDLY107, SDLY1, SDLY107RV, and SDLY107(1–5′UTR)RV strains were grown at 37 ℃ (C) or 39.5 ℃ (D) in RD cells with initial inoculation at a multiplicity of infection of 100. Data show the means ± SDs of three samples. Each experiment was carried out in triplicate. Error bars represent SDs.
The infectious RNAs were transcribed in vitro from linearized pcDNA3.1(+)SDLY107 and pcDNA3.1(+)SD LY107(1–5′UTR)using a Large Scale RNA Production System-T7 kit(Promega, USA)according to the manufacturer's instructions. After purification(MicroElute RNA Clean-Up Kit; OMEGA, USA), the synthesized RNA transcripts were transfected into RD cells using TurboFect Transfection Reagent(Thermo, USA). At 24 h post-transfection, RD cells transfected with the rescued SDLY107RV or SDLY107(1–5′UTR)RV RNA showed typical cytopathic effects(CPE), such as cell rounding, aggregation, detachment, and floatation(Figure 1B). In addition, the CPE developed at same rates in the transfected cells. The rescued viruses were harvested after three consecutive freeze-thaw cycles and stored at -80 ℃ after three passages. The viral titers of the rescued SDLY107RV and SDLY107(1–5′UTR)RV strains were similar to that of the wild-type strain(106.5 median tissue culture infective dose [TCID50]/mL). These results demonstrated that RNA transcripts from the cDNAs were infectious and that the rescue of EV71 was successful.
To illustrate the kinetics of viral replication, the wildtype strains SDLY107 and SDLY1 and the rescued stains SDLY107RV and SDLY107(1–5′UTR)RV were harvested from RD cells at 12-h intervals, and the levels of viral RNAs were measured by quantitative real-time PCR. The growth curves of viruses in RD cells at a multiplicity of infection(MOI)of 100 are shown in Figure 1C and Figure 1D.
At 60 and 84 h after inoculation, the viral RNAs of all three strains peaked and were then maintained between 84 and 96 h, with the exception of the SDLY1 strain, in which RNAs decreased from 72 to 96 h after inoculation at 37 ℃. At 37 ℃, similar proliferation rates were observed up to 96 h after inoculation between the rescued and wild-type EV71 strains. However, the maximum viral load of the rescued strains was slightly lower than that of the wild-type strain. Additionally, the growth of all four strains was inhibited to temperatures above the optimal temperature range(37 ℃ versus 39.5 ℃), particularly for the recombinant rescued SDLY107(1–5′UTR)RV strains and the SDLY1 strain. The levels of viral RNAs of SDLY107(1–5′UTR)RV and SDLY1 were approximately 10-fold lower than those of SDLY107 and SDLY107RV from 24 to 96 h after inoculation at 39.5 ℃. The growth curves demonstrated that viral replication occurred in the transfected RD cells and confirmed the success of EV71 rescue. The replication capacity of SDLY 107(1–5′UTR)RV was different from that of SDLY107 at the higher temperature(39.5 ℃)compared with that at the optimal temperature(37 ℃), suggesting that the different replication capacities among the four strains at the higher temperature were influenced by the 5′UTR and that the 5′UTR may contribute to the temperature tolerance of the strains, to some extent.
For many years, researchers worldwide have attempted to elucidate the pathogenic mechanisms of EV71. A number of studies have demonstrated that antibody-dependent enhancement(ADE)infection and variations in VP1 and 3C protease contribute to the severity of EV71 infection(Chen et al., 2013; Wang et al., 2010; Zhang et al., 2014; Lei et al., 2010). In contrast, few reports have described the importance of the 5′UTR in EV71 virulence. As mentioned above, the stem-loop Ⅰ element at the N-terminus of the 5′UTR plays an important role in viral RNA replication, and the IRES can initiate genome translation through an IRES-dependent translation mechanism(Luo et al., 2014). Yeh et al.(2011)showed that a single nucleotide change(C158U)in stem-loop Ⅱ attenuates EV71 virulence. During replication and translation mediated by the 5′UTR in EV71, it is fundamentally important for viruses to interact with cellular proteins, such as poly(C)-binding protein 1(Luo et al., 2014). Interestingly, the difference in viral replication between SDLY107(1–5′UTR)RV and SDLY107 was observed only at the higher temperature(39.5 ℃), suggesting that the different replication capacities among the four strains at the high temperature were influenced by the 5′UTR and that the 5′UTR may contribute to the temperature tolerance to some extent. An alternative explanation for this replication difference is that the structure of the proteins encoded by the viruses may be altered as the temperature changes. However, this hypothesis needs to be studied further.
In a previous study, we found that high and low virulence EV71 isolates exhibit distinct replication capacities and that EV71 replication is a key virulence factor(Sun et al., 2014). Thus, because of the replacement of the 5′UTR of SDLY107, the virulence transformed from high to low, corresponding to the variation in replication capacity. While more research need to be carried out to further confirm this statement such as neurovirulence experiments tested on murine.
In this report, we presented the construction of an infectious full-length clone and a recombined EV71 cDNA clone, from which the RNA transcripts could result in CPE in transfected cells. Furthermore, we also found that the 5′UTR of EV71 could influence the replication of EV71, potentially acting as a determinant of the virulence of EV71. Our results provide important insights into the pathogenic mechanisms of EV71-related diseases.
-
This study was supported by National Natural Science Foundation of China(81371833) and Medical and Health Science and Technology Development Plan of Sh and ong Province(2013WS0211). The authors declare that they have no conflict of interest. This study was approved by the ethical committees of School of Public Health, Sh and ong University, Jinan, Sh and ong 250012, China(permit number 20080301). Written consents were obtained from all children's parents involved in the study.
Supplementary Table S1 is available on the website of Virologica Sinica: www.virosin.org; link.springer.com/journal/12250.
-

Table S1. Primers for the construction of the infectious cDNA clone and recombinant viruses.
















 DownLoad:
DownLoad: