HTML
-
Avian Influenza virus (AIV) H9N2 subtype viruses circulate widely in domestic fowl, and usually cause mild clinical signs in poultry (Li et al., 2005) . Occasionally, avian H9N2 can infect humans and cause mild clinical symptoms (Peiris et al., 1999; Lin et al., 2000) . Genetic analysis indicates that the H9N2 genotype viruses exist in major poultry species (such as duck and chicken) and in minor poultry species (such as quail, partridge, chukar, pheasant, and guinea fowl) (Guan et al., 2000; Li et al., 2005) . Meanwhile, frequent reassortment events among different H9N2 genotypes and between H9N2 and other AIV subtypes have been detected (Deng et al., 2013; Chen et al., 2014) . These situations raised the public health concern that H9N2, as well as H5N1, are the most likely candidates to cause a new influenza pandemic in humans (Li et al., 2005; Liu et al., 2014) .
From Nov 2013 to Feb 2014, three human cases of a novel influenza A virus (H10N8) infection were reported in China (Chen et al., 2014) . Preliminary virus isolation and genetic analysis suggested that the novel human-infectingin fluenza virus (H10N8) was genetically distinct from the avian influenza A (H10N8) viruses previously identified in China. One hypothesis is that the emerging H10N8 virus resulted from multiple reassortments of subtype H9N2 strains in poultry in China (Chen et al., 2014; Yang et al., 2014) . A recent study of viral culture in chicken eggs and in MDCK cells also suggests that this virus originated from fowl (Yang et al., 2014) .
In this report, we identified a novel strain of influenza A (H9N2) virus in chicken in Hubei, China. Genomic and phylogenetic analysis indicate that the hemagglutinin (HA) and neuraminidase (NA) genes of this H9N2 are similar to AIV H9 and N2 subtypes, but six internal genes (PB1, polymerase basic protein 1; PB2, polymerasebasic protein 2; PA, polymerase acidic protein; NP, nucleoprotein; M, matrix; NS, nonstructural protein.) are homologous to the emerging human H10N8 virus.This data provides further genetic evidence that the human H10N8 virus originated from avian H9N2 viruses, via reassortment events.
In May 2014, we obtained ten swab samples (a cloacal swab and tracheal swab from the same chicken were put into the same sample collection tube and counted as one sample) from a brood of chickens showing mild clinical signs in a farm. Swab samples were kept in virus transportmedium (VTM) buffer containing penicillin (100 U/mL) and streptomycin (100 μg/mL) . Viral RNA (vRNA) was extracted from samples using QIAmp Viral RNA Minikit (Qiagen, Hilden, Germany) . A set of pan-PCR primers (FluMU44 and FluML287, Table 1) for influenza A virus against M gene were used to test the samples. RTPCRwas performed in a 25 μL reaction using the One-Step RT-PCR kit (Invitrogen, Van Allen Way Carlsbad, CA) . From all the RNA extracts of ten samples (Supplementary Figure S1 lanes 1–10) , the 244 bp products at the expected size were successfully amplified. The bands were further gel purified with a DNA extraction kit (E.Z.N.A.® Gel Extraction Kit, OMEGA, Norcross, GA) and directly sequenced with a 3700 DNA Sequencer (ABI, Foster City, CA) . Sequencing result confirmed that all the samples were AIV positive and the ten partial M genes were complete identical (data not shown) .
Primer name Oligo-sequences FluMU44 5′-GTCTTCTAACCGAGGTCGAAACG-3′ FluML287 5′-GCATTTTGGACAAAGCGTCTACG-3′ Table 1. Influenza A virus testing primers.
To further characterize this virus, one sample was used to determine the full-length viral genome by reverse transcription (RT) with the Uni12 primer and amplifying all the genes using eight sets of segment-specific primers complementary to the conserved nucleotides at the 5′ and 3′-end of the vRNA (Phipps et al., 2004) . Eight fulllength fragments were successfully amplified from the sample (Supplementary Figure S2) . Raw sequences of each gene segment were analyzed using Geneious software (Version 5.5.9) to assemble to full-length genome, and further gene alignment results showed that this virus belonged to influenza A H9N2 subtype and was named as A/chicken/Hubei/01/2014 (H9N2) . The genomic length of this H9N2 virus was 13, 596 nucleotides (nt) long, and the PB1 and PB2 each segment consists of 2, 341 nt, the PA, HA, NP, NA, M, and NS each segment consists of 2, 233, 1, 742, 1, 565, 1, 457, 1, 027, 890 nt, respectively.The complete genome sequences of this H9N2 strain were deposited under accession Nos. KT164848–KT164855 in GenBank.
By comparing sequences, we found that the nucleotide and the coded amino acid sequences of eight genes of A/chicken/Hubei/01/2014 (H9N2) showed 97.2%–99.7% and 96.4%–100% identities compared to previously identified influenza viruses, respectively.The HA and NA genes were most close and shared 98.6% and 97.2% nucleotide sequences identity to HA and NA genes of H9N2 subtype viruses in GenBank (A/chicken/Zhejiang/C497/2013 (H9N2) , A/chicken/Shanghai/C1/2012 (H9N2) , etc) which were also found in chicken in different provinces of China (Feng et al., 2013) . The most interesting finding is that the 6 internalgenes of A/chicken/Hubei/01/2014 (H9N2) were high homologous (shared 99.3%–99.7% nucleotide sequence identities) to the corresponding genes of theemerging human-infecting H10N8 viruses (A/Jiangxi/IPB13a/2013 (H10N8) , A/Jiangxi/IPB13/2013 (H10N8) , A/Jiangxi/IPB13c/2013 (H10N8) , and A/Jiangxi/IPB-13b/2013 (H10N8) ) , which were previously reported in Jiangxi province, in China (Chen et al., 2014; Yang et al., 2014) .
To better underst and viral evolutionary status, the coding sequences of each segment of A/chicken/Hubei/01/2014 (H9N2) were aligned with related viruses in GenBank. The phylogenetic relationships were calculated by the neighbor-joining method, based on the sequence alignments of every gene generated by Geneiouss oftware. Nucleotide substitutions were set under the Kimura 2-parameter model, and substitution rates among sites were set in gamma distribution. Bootstrap values were tested from 1, 000 replicates. The lengths of complete nucleotide coding sequence used for the phylogenetic analysis were as follows: PB1 2, 274 nt, PB22, 210 nt, PA 2, 151 nt, NS 838 nt, NP 1, 497 nt, NA 1, 398 nt, M 982 nt, HA 1, 686 nt. In the phylogenetic tree, the HA and NA genes of A/chicken/Hubei/01/2014 (H9N2) clearly fell into the H9 and N2 clades respectively. But, the six internal genes formed the distinguished brancheswith human sourced H10N8 viruses (Figure 1) . In addition, the HA of human H10N8 viruses clustered with known H10 subtype viruses found in duck or mallard (A/duck/Hunan/S11205/2012 (H10N3) and A/mallard/Sweden/105404/2009 (H10N1) ) . The NA of human H10N8 viruses clustered with known N8 subtype viruses also found in avian (A/duck/Guangxi/GXd-6/2010 (H6N8) ) (Figure 1) . The high similarities and close evolutionary relationships of internal genes between H9N2 and H10N8 strongly suggested that the emerging human H10N8 virus obtained the internal genes from chicken H9N2 viruses.
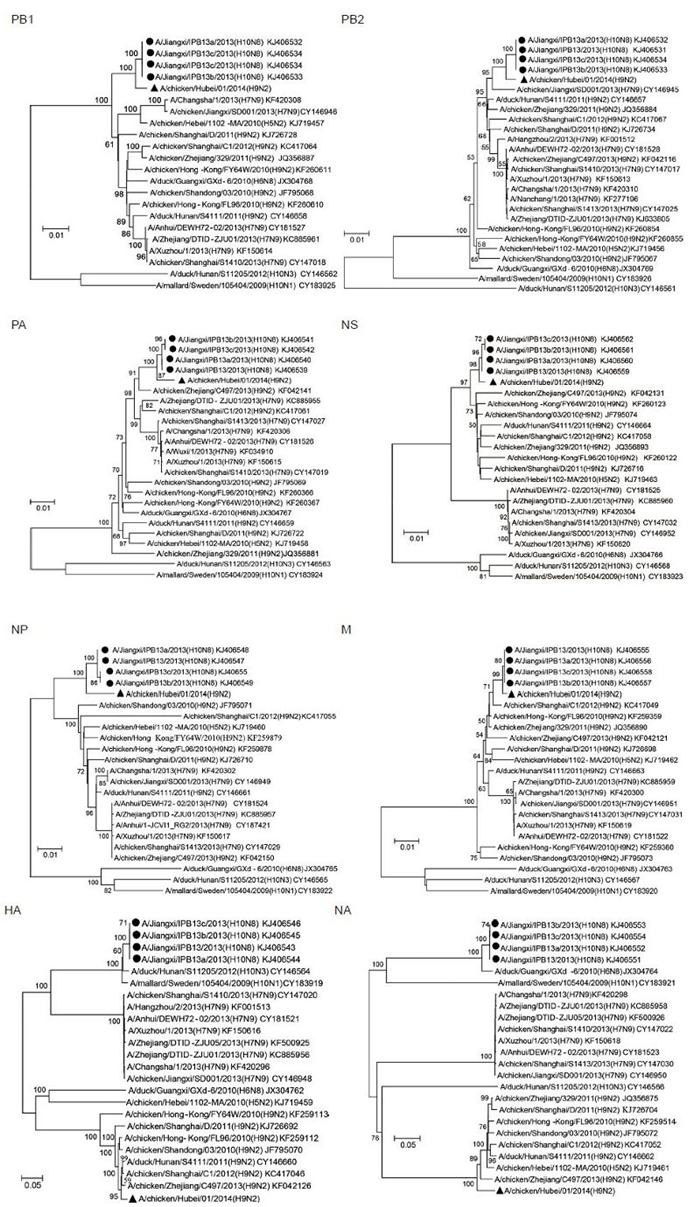
Figure 1. Phylogenetic analysis of the PB1, PB2, PA, HA, NP, NA, M, NS genes of the H9N2 influenza virus with related influenza A viruses in GenBank. The phylogenetic relationships were calculated by the neighbor-joining method. Bootstrap values were tested from 1000 replicates, tested values > 50% were shown in the nodes. Viral full names and GenBank accession numbers were shown in the tree. The A/chicken/Hubei/01/2014 (H9N2) virus obtained in this study was labeled by triangle, H10N8 virus strains derived from human samples were labeled by circle.
From known results, two possible reassortment routes of the human H10N8 virus could be speculated. One route involves the H9N2 virus providing the six internal genes directly to a H10N8 virus; this reassortment event could generate the precursor of human H10N8 virus through one step. The other route is the H9N2 virus went through two-step reassortment events, first with a H10 subtype (like H10N3) virus, then a N8 (like H6N8) subtypevirus (or in the reverse order) , to generate the precursor of human H10N8 virus. The reassortment H10N8precursor virus was circulating in domestic poultry, occasionally infecting humans.
Recently, phylogenetic analyses suggest that different subtypes of AIV (H7N9, H6N1, H10N8) co-circulated with H9N2 and have reassorted their internal genes (Shi et al., 2014) . AIV in poultry surveillance in southern China indicates that H9N2 virus distributes widely (Liu et al., 2014; Yu et al., 2014) . Meanwhile, it is assumed that live poultry markets play an important role in the ecology of AIV (Liu et al., 2003; Kim et al., 2006; Jadhao et al., 2009; Liu et al., 2014) . Considering above causes, further extensive surveillance of AIV in poultry and wild fowl may help to predict the potential interspecies transmission risk.
We identified a novel strain of influezna A H9N2 virus infection in chicken in China. Genomic and phylogenetic investigation revealed that the internal genes of the novel H9N2 virus strain were closely related to the emerging human H10N8 virus. These results suggest that the chicken H9N2 virus may be the internal gene source of the novel human H10N8, and through one or two step reassortment events could generate the precursor of the novel human H10N8. This data provides further evidence to address the hypothesis of avian origin of the novel human H10N8. However, the reassortment processes of the poultry H9N2 virus with other subtype influenza A viruses have not been characterized. More research of AIV surveillance and genetic analysis is required to better underst and the origin of the emerging influenza virus.
-
We acknowledge financial support from the Youth Innovation Promotion Association of Chinese Academy of Sciences (2014309) . We thank Adam Nitido for language editing. The authorsdeclare that they have no conflict of interest. This article doesnot contain any studies with human or animal subjects performed by any of the authors.
Supplementary figures are available on the website of Virologica Sinica: www.virosin.org; link.springer.com/journal/12250.
-
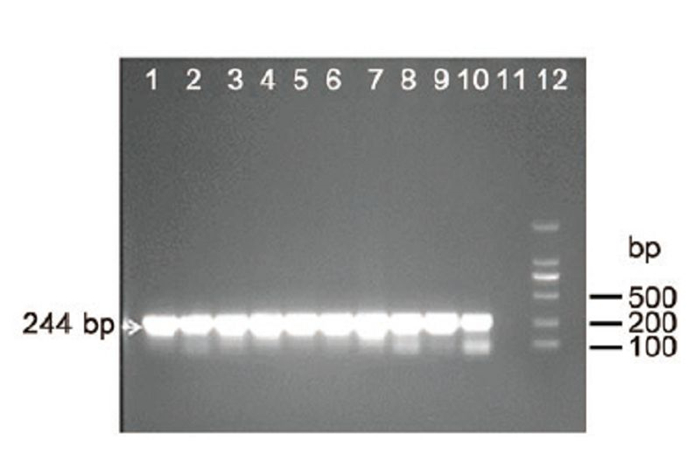
Figure S1. RT-PCR detection of influenza A virus using pan-influenza A primers. Lanes 1–10: RT-PCR amplification of viral RNA in ten samples with primers FluMU44 and FluML287; Lane 11: negative control; Lane 12: DNA ladder 2000. Target 244 bp bands were indicated by arrow. Lanes 1–10: 2 μL of total RNA extracted from swabs were added to the RT-PCR reaction. Lane 11: 2 μL H2O was added instead of 2 μL RNA to the PCR reaction.
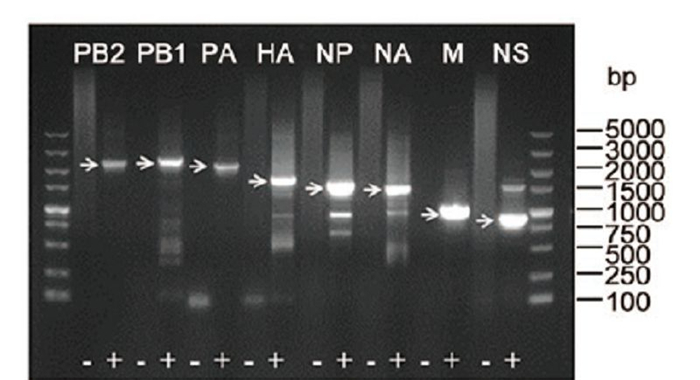
Figure S2. Full length amplification of all eight segments of A/chicken/Hubei/012014 (H9N2) by RT-PCR. Reverse transcription of total RNA extracted from swab was performed with Uni12 primer. Subsequently, PCR reactions for each segment were performed using the eight sets of segment-specific primers. Target bands were indicated by arrows. As a negative control (−) H2O was added instead of reverse transcription product (+) to the PCR reaction.
















 DownLoad:
DownLoad: