-
Dear Editor,
Astrovirus is an enteric virus associated with sporadic diarrhea or large outbreaks of gastroenteritis (Mendez et al., 2013). Until 2008, human astroviruses (HAstVs) were classified into eight serotypes—HAstV-1 to HAstV-8—based on the reactivity of their capsid protein with type-specific antibodies. Among these serotypes, HAstV-1 is the most prevalent globally. However, some divergent new astroviruses (MLB1/MLB2, VA1/VA2/VA3, and HMO A/B/C) have been discovered recently in stool samples of patients with and without diarrhea (De Benedictis et al., 2011).
HAstVs are endemic pathogens, and outbreaks of HAstV-induced gastroenteritis have been frequently reported worldwide. However, reports on gastroenteritis outbreaks in childcare centers remain limited. We report here an HAstV outbreak in a childcare center in China. In November 2010, 35 three-to four-year-old children from the same childcare center in a community of Jinzhou City, Liaoning Province were diagnosed with acute diarrhea. All 35 children presented with water diarrhea. No red cells or pyocytes were observed in routine stool examination. Fever was observed in 10 patients (temperature range, 37.5 °C to 38.3 °C; 7 male and 3 female). No vomiting or abdominal pain was observed in any of the patients. The mean frequency of loose stools was 3 times per day, and the mean duration of diarrhea was 2.5 days. All children were hospitalized to receive fluid and electrolyte replacement. Resolution was good in all patients. To determine the etiological agent, stool samples were collected from these 35 children and 40 healthy children (negative control). The fecal suspensions were screened for group A rotavirus, adenovirus, and astrovirus using ELISA kits (Dako Diagnostics, Denmark). Viral RNA was extracted from each stool suspension using TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Subsequently, RT-PCR was performed to detect calicivirus (norovirus GI, GII, and sapovirus), adenovirus, and astrovirus according to previously published protocols (Walter et al., 2001; Phan et al., 2005).
All 35 patient stool samples were negative for rotavirus, adenovirus, and calicivirus. However, HAstV was detected in all samples by RT-PCR and in 30 of 35 samples (85.7%) by ELISA. The control group samples were negative for rotavirus, adenovirus, calicivirus, and astrovirus. All 35 isolates from the patients were sequenced, and 15 newly identified strains were deposited in GenBank(accession number: KF211460-KF211474). HAstV ORF2 encodes a capsid precursor and harbors a 348-bp region that is known to be conserved across strains and lineages. BLAST analysis of the348-bp region of the ORF2 of these 15 HAstV stains revealed 98%-99% homology to the corresponding sequence in HAstV-1. To study the genetic relationship between these 15 astrovirus stains and those reported previously, a phylogenetic tree was constructed based on the sequence of the 348-bp ORF2 segment (amplified using the Mon 269 and Mon 270 primers). HAstV-1 is classified into six lineages (HAstV-1a-f) (Gabbay et al., 2007), and all sequences in this study clustered under lineage 1d (Figure 1A), with 89.08%-98.56% sequence homology.
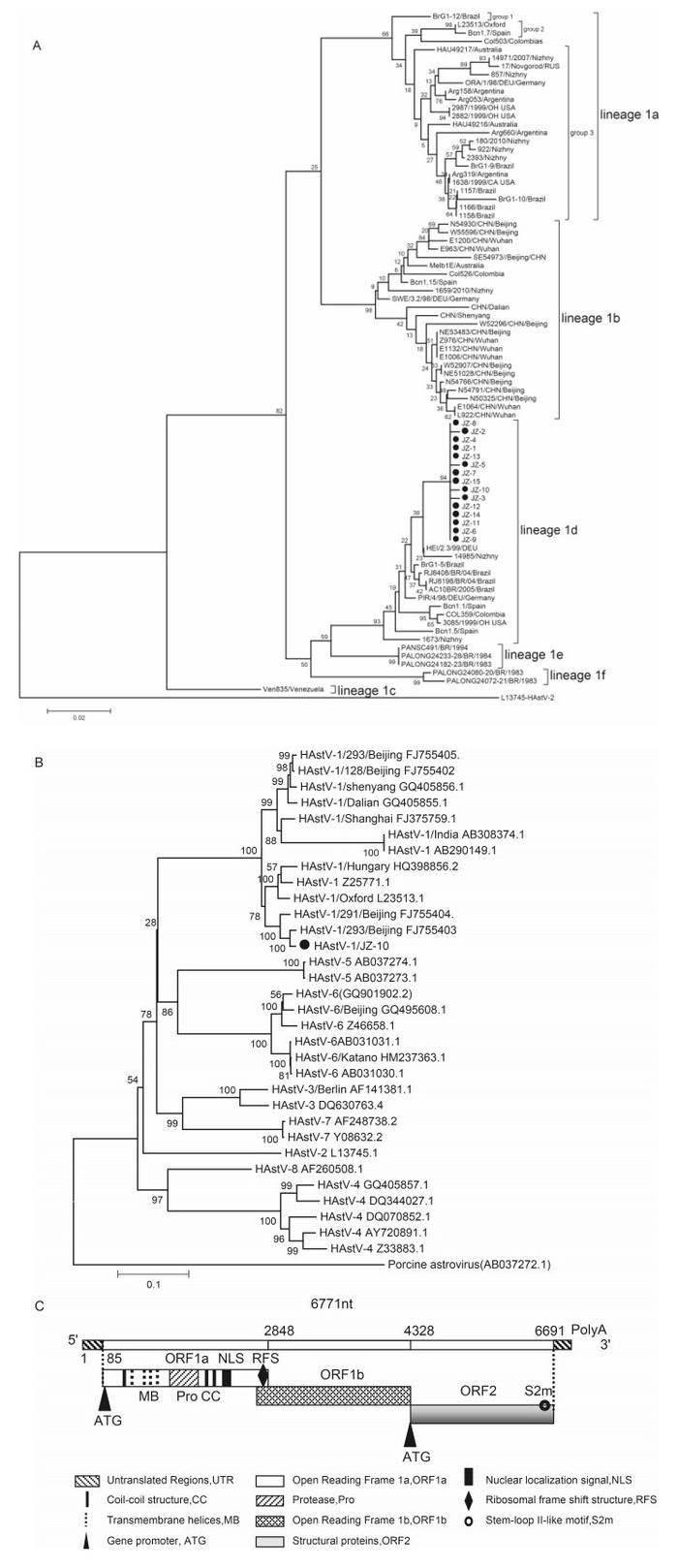
Figure 1. (A) Neighbor-joining phylogenetic tree of the capsid precursor-encoding 348-bp region of ORF2 of the human astrovirus (HAstV) strains isolated from a childcare center in Jinzhou City, China. (Collection date, 18 November 2010; denoted by ●). The bar on the left indicates the genetic distance. Internal labels represent the bootstrap values of 1000 replicates. (B) Neighbor-joining phylogenetic tree based on the sequence of ORF2 of the 32 strains of HAstV-1/JZ (labeled by ●); porcine astrovirus sequence was the out group. (C) Molecular characterization of the genomic RNA of HAstV-1d JZ strain (GenBank ID: KF211475). The genome organization with the predicted amino acid motifs is shown. The locations of the 3 ORFs, gene promoter (ATG), non-coding region (NCR), predicted transmembrane helices (MB), protease (pro), nuclear localization signal (NLS), coil-coil structure (CC), ribosomal frameshift structure (RFS), and stem-loop Ⅱ-like motif (s2m) are indicated.
An HAstV-1-positive stool sample, JZ-10, was used to synthesize the full-length viral genome sequence. Eleven overlapping cDNAs, accounting for the complete HAstV-1 genomic RNA, were synthesized by RT-PCR. Supplementary Table S1 shows the primers sequences and their positions within the HAstV-1 genome. Both cDNA synthesis and RT-PCR were performed using standard protocol (Jiang et al., 2013).The genomic RNA of the HAstV-1d JZ-10 stain comprises 6771 nucleotides (nt), including a 5′-untranslated region (5′-UTR; 85 nt), 3 ORFs, a 3′-UTR (80 nt), and a poly-(A) tail at the 3′ end (Figure 1C). The protein-coding region of 6609 nt was organized into 3 ORFs, similar to that of other astroviruses (Figure 1C). The full-length genomic RNA showed 91.4%, 90.1%, and 89% sequence homology to the genomes of HAstV-1/Oxford (L23513), HAstV-1/Beijing (FJ755402), and HAstV-1/Shanghai (FJ375759) strains, respectively. In the phylogenetic analysis based on the complete capsid precursor protein of the JZ-10 stain and related astrovirus strains available in GenBank, the JZ-10 stain clustered to lineage HAstV-1d (Figure 1B). The full-length genomic sequence was deposited in the GenBank (ID: KF211475).
Epidemiological studies have reported that in China, the overall incidence of astrovirus infections in young children hospitalized with diarrhea is 2%-9% and that HAstV-1 infection-associated diarrhea is widespread among hospitalized children, with type 1 being the predominant serotype. Most of this information about is based on hospitalized diarrhea patients and those with coinfections with rotavirus, norovirus, and adenovirus (Guo et al., 2009; Jin et al., 2013; Ren et al., 2013). To the best of our knowledge, no HAstV-1 outbreaks have been reported in Chinese childcare centers. Moreover, most astrovirus outbreak reports do not mention the lineage. Molecular epidemiological studies revealed that the astrovirus infection in infants in Wuhan, Beijing, Shanghai, Dalian, Shenyang, and Inner Mongolia were all associated with HAstV-1b lineage (Liu et al., 2004; Shan et al., 2009; Dai et al., 2010; Li et al., 2010; Wang et al., 2011). However, in this study, all strains were clustered under lineage 1d. Additionally, although Jinzhou city shared the same geographic location and province with the cities of Shenyang and Dalian, the lineage of the isolates was different.
We confirmed gastroenteritis outbreak cause by astrovirus lineage 1d in a Chinese childcare center. We did not detect other intestinal viral infections and new astroviruses, such as MLB1, AV, and HMOAstV. Owing to the availability of more sensitive and specific diagnostic tests, HAstV has emerged as a predominant cause of diarrhea and has been associated with food-and water-borne illnesses. In this study, we did not analyze samples environmental samples from the childcare center. Therefore, the relationship between the infection and persistence of HAstV-1d in the environment was not established. Nonetheless, we speculate that poor hygiene in the childcare center likely contributed to the spread and persistence of HAstV. Although the diarrhea induced by HAstV-1d in this childcare center was mild, the disease disappeared in an average of 1 day after hospitalization. The role of astrovirus in infections reported in childcare centers should not be neglected and hygiene practices in childcare rooms should be improved to prevent further outbreaks.
To our knowledge, this study is the first to link a specific HAstV serotype (HAstV-1d) to an outbreak of diarrhea in a childcare center. Our results will be useful in improving astrovirus surveillance and control measures. Longitudinal studies are needed to investigate the role of HAstV in diarrhea in children in the Liaoning Province.
HTML
-
This work was supported by the National Nature Science Foundation of China (No. 81201285), Dr. Start-up Fund of Liaoning Province (No. 20141134), excellent talents project in colleges and universities of the Liaoning Province Foundation (No. LJQ2015067), college students of science and technology innovation fund, Liaoning medical university principal fund (No. 2014D09). The authors declare that they have no conflict of interest. The study was approved by the ethics committees of Liao Yi [2014] (NO.6-20150005). All participants provided written informed consent. Written consents were obtained from all children's parents involved in the study.
Supplementary Table S1is available on the website of Virologica Sinica: www.virosin.org; link.springer.com/journal/12250.
-
Name Primer sequence(5′-3′) Annealing temperature (Tm, ℃) Product(bp) Primer location* JZ-P1 F ACCTGACAAAGATTTCAACTTTGGG 55 807 117–923 R GACAAGTGGGACGTTGCAAGTGTAA JZ-P2 F CCAAGAGGGGGGTGATTGGCCT 55 312 1–312 R TCCATCCTCAAGTCCACCAGTAATC JZ-P3 F GCAAATATAACCATCAGTGACAAGC 53 804 712–1515 R CATCAAGCCCTCATAGCACACATTC JZ-P4 F TTCGTTGTTATAAAACCAGGTGCGT- 54 709 1369–2077 R TTTGTAAGAATGGTGCAAGAATCCC JZ-P5 F AGAACTCGACGACTGTCGTCACACA 50 719 1919–2637 R TGCTTTAGTTAGTCGGTGTGGCGCT JZ-P6 F AGGCACCAATCTGGGAATCTTATGA 50 768 2549–3316 R GTGAATTCTCTTATGTATGGTGCCC JZ-P7 F TGGGTGGGCACCATACATAAGAGAA 50 822 3287–4108 R TTTTCTGGAGAAGTTGGCACGGGTT JZ-P8 F GTGGGTTAAGCCTGGGAAGGTCATC 53 812 4001–4812 R AGTACCATTGACAGCAGATGCGCCA JZ-P9 F GGCGTTAGGTGCACAATATTCCATG 52 665 4722–5386 R CCTTGACTAACCAGTTGAACGGCGG JZ-P10 F CAGACACAGTATGGCAGGTGCTCAA 51 728 5303–6030 R AGTGGCAGACGCCCTGTAAAAGT JZ-P11 F TATAGATTTTCAACCATCCGAGGCG 55 817 5817–6633 R ATGAGACCATGCACTGTTTGGGCTT 3'-GSP1 ATTCACCTGATAATGCCTTTGCC 54 400 6396–terminal 3'-GSP2 AGAGGACGAAGACGACGAAGCG 50 Note: *HAstV-1 Oxford L23513.1 Table Table S1. Primer sequences used for RT-PCR analysis of complete genome of HAstV-1 JZ strain
















 DownLoad:
DownLoad: