HTML
-
Baculoviridae is a family of rod-shaped, invertebrate-specific DNA viruses with double-stranded, covalently closed, circular genomes ranging from 80-180 kb in size (Lange and Jehle, 2003). Based on molecular phylogenetic analysis, the family is divided into four genera: lepidopteran-specific nucleopolyhedroviruses (NPVs) and granuloviruses are classified as an Alphabaculovirus and a Betabaculovirus, respectively, and NPVs that infect hymenopteran and dipteran insects are classified as a Gammabaculovirus and a Deltabaculovirus, respectively (Jehle et al., 2006). The alphabaculoviruses are further divided into Group Ⅰ and Group Ⅱ based on phylogenetic analyses (Hayakawa et al., 2000; Herniou et al., 2001; Herniou et al., 2003). Most baculoviruses undergo a biphasic life cycle that generates two virion phenotypes: the occlusion-derived virus (ODV) transmits infection from insect to insect (oral infection), while the budded virus (BV) mediates systemic infection within the infected insect (Funk et al., 1997). Upon ingestion via contaminated food, the occlusion bodies (OBs) dissolve in the alkaline environment of the larval midgut and release the ODVs, which infect midgut epithelial cells by membrane fusion (Federici, 1997). The tracheae serves as a conduit for progeny virions to cross the basal lamina (BL) into the hemocoel of the insect and establish systemic infection within the insect body (Engelhard et al., 1994).
Fibroblast growth factors (FGFs) are members of a large family of growth factors that are widespread in vertebrates and invertebrates. In vertebrates, 22 members have been identified in the FGF family, sharing highly conserved gene structure and amino acid sequence (Itoh and Ornitz, 2004). FGFs have an array of diverse functions in both developing and adult tissues, including regulating cell proliferation, migration, and differentiation, as well as tissue repair and injury response (Ornitz and Itoh, 2001; Sato and Kornberg, 2002). Although widespread in multicellular organisms, fgf homologs have only been identified among viruses in baculoviruses (Detvisitsakun et al., 2007). The lepidopteran baculovirus viral fibroblast growth factors (vFGFs) facilitate dissemination of the virus from infected midgut epithelial cells (Detvisitsakun et al., 2007).
The function of vFGF in Group Ⅰ alphabaculovirus, including Autographa californica multiple nucleopolyhedrovirus (AcMNPV) and Bombyx mori nucleopolyhedrovirus (BmNPV), has been studied. Baculovirus vFGFs share a number of properties common to multicellular-organism FGFs, including structural features, extracellular secretion, heparin affinity, and stimulation of insect-cell motility (Katsuma et al., 2004; Detvisitsakun et al., 2005). Deletion of vfgf in AcMNPV exhibited no obvious effects on infectious BV production in cultured cells (Detvisitsakun et al., 2006); however, vfgf deletion delayed the time of death in Spodoptera frugiperda and Trichoplusiani larvae when the virus was tested by feeding, but not by intrahemocoelic injection (Detvisitsakun et al., 2007). In contrast to AcMNPV, the BmNPV vfgf-knockout mutant produced fewer BVs in cultured cells, but it also took longer to kill the B. mori larvae as compared to the wild-type virus when delivered either by feeding or by intrahemocoelic injection (Katsuma et al., 2006b). The roles of vFGF in systemic infection establishment involve remodeling of the BL, which is a tightly woven, virus-impenetrable lining located between the midgut epithelial cells and tracheal cells, and stimulation of tracheoblast migration toward vFGF-expressing cells (Means and Passarelli, 2010). The tracheal system then mediates viral escape from the midgut, carrying the virus to hemocoelic cells and disseminating the infection to other tissues of the infected insects.
Given the functional discrepancies between the two reported vFGFs, it is of interest to investigate the functions of vfgf from different baculoviruses. The vFGF of Group Ⅱ alphabaculoviruses is different from that of Group Ⅰ alphabaculoviruses, with a larger C-terminal region and more N-linked glycosylation consensus sequences (0-4 for Group Ⅰ; 3-12 for Group Ⅱ) that may contribute to vFGF secretion (Katsuma et al., 2004; Katsuma et al., 2006a). In this study, we investigated the function of vFGF from the Group Ⅱ alphabaculovirus Helicoverpa armigera single nucleopolyhedrovirus (HearNPV). In order to elucidate vFGF roles during HearNPV infection in vitro and in vivo, we characterized vfgf transcription and expression profiles and constructed vfgf-knockout and -repair HearNPV recombinants. The results suggested that HearNPV vfgf play roles in the mortality of host as that of Group Ⅰ alphabaculoviruses.
-
HzAM1 cells were maintained in Grace's insect medium (Gibco-BRL; Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Life Technologies) at 28℃. vHaBac-egfp-ph was used as the wild-type control virus (Song et al., 2008) and bHaBacHZ8, an infectious HearNPV bacmid, was used as the parental bacmid (Wang et al., 2003). H. armigera larvae were reared on an artificial diet at 27℃.
-
To generate anti-vFGF antibodies, a truncated vfgf fragment (70-906 nt) was amplified with the primers anti-vFGF-f and anti-vFGF-r (Supplementary Table S1) using bHaBacHZ8 DNA as the template. The fragment was then cloned into a pET-28a expression vector (Novagen; Merck Millipore, Billerica, MA, USA) and expressed as a His-tag fusion protein in Escherichia coli BL21 cells. The fusion protein was purified and used to generate rabbit polyclonal antiserum (anti-vFGF) according to a previously reported method (Wang et al., 2008).
To analyze the transcription and expression profile of HearNPV vfgf, 5×105 HzAM1 cells were infected with vHaBac-egfp-ph at a multiplicity of infection (MOI) of 10 50% tissue-culture infection dose units/cell. At 0, 3, 6, 12, 24, 36, 48, and 72 h post-infection (h.p.i.), cells were collected and washed with phosphate-buffered saline (Figure 1B and 1C). For transcriptional analysis, total DNA-free RNA harvested from the infected cells at each time point was isolated by Trizol reagent (Invitrogen, Carlsbad, CA, USA), and the first-strand cDNA was synthesized with oligo (dT) primers (Promega, Madison, WI, USA) using M-MLV reverse transcriptase (Promega, Madison, WI, USA). The cDNA was then amplified with primers vFGF-trans-f and vFGF-trans-r (Supplementary Table S1). After DNase Ⅰ treatment, RNA from each sample was also tested by PCR for DNA contamination.
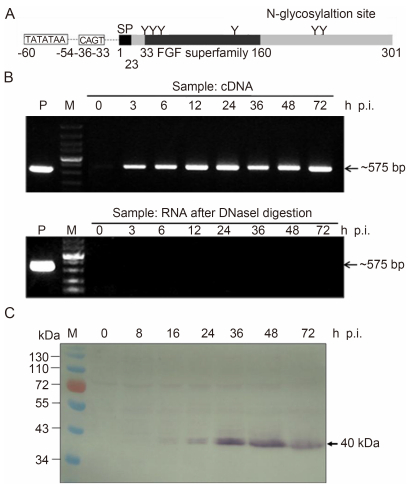
Figure 1. Transcription and expression analyses of vFGF. (A) Schematic representation of HearNPV vFGF. The relative positions of the predicted signal peptide (SP), N-glycosylation sites, and early promoter are shown. (B) Time-course analysis of vFGF transcription. HzAM1 cells were infected with HearNPV BV at an MOI of 10 and collected at the indicated post-infection time points. Total cellular RNA after DNaseI treatment was determined by RT-PCR (upper panel). RNA in each sample after treatment with DNaseI was assessed for DNA contamination by PCR (lower panel). Total DNA in infected cells collected at 72 h.p.i. was used as a positive control. (C) Time-course analysis of vFGF expression. HzAM1 cells were infected with vHaBac-egfp-ph at an MOI of 10 and collected at the indicated post-infection time points. Cellular proteins were separated by 10% SDS-PAGE, and the western blot was probed with anti-vFGF polyclonal antibody. The vfgf bands are indicated by arrows. M, DNA or protein molecular marker.
For expression analysis, protein samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; 10% acrylamide gel) and electroblotted onto a nitrocellulose membrane. The blot was probed with anti-vFGF as the first antibody and goat anti-rabbit IgG conjugated with alkaline phosphatase as the secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA). The final signals were detected with 5-Bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium.
-
The vfgf gene from bHaBacHZ8 was knocked out using a modified form of a previously reported method (Hou et al., 2002). The flanking sequences of the vfgf gene were PCR amplified using bHaBacHZ8 as a template and the primers del-F1f/del-F2r and del-F2f/del-F2r (Supplementary Table S1). PCR products were confirmed by sequencing and cloned into the pKS-egfp-cat plasmid (kindly provided by Prof. J.M. Vlak, Wageningen University), which contains the enhanced green fluorescent protein (egfp) gene under control of the HSP70 promoter and the chloramphenicol resistance gene (cat) (Luo et al., 2011). The resulting plasmid was designated pKS-F1-egfp-cat-F2. The fragment containing the egfp and cat genes flanked by F1 and F2 was then excised from pKS-F1-egfp-cat-F2 using Kpn Ⅰ and Xba Ⅰ restriction enzymes to obtain a 3.3-kb fragment, which was gel purified and transformed into E. coli BW25113 electrocompetent cells containing bHaBacHZ8 and the helper plasmid pKD46 to generate bHaBac-Δvfgf-egfp (Figure 2A) (Hou et al., 2002). Positive clones resistant to both kanamycin and chloramphenicol were confirmed by PCR.

Figure 2. Construction and verification of recombinant HearNPVs. (A) Construction of the recombinant HearNPV bacmids. The fragment containing the egfp and cat genes flanked by vfgf homologous arms was used for homologous recombination to generate bHaBac-Δvfgf-egfp. The ph gene was reintroduced into bHaBac-Δvfgf-egfp by Tn7-mediated transposition to produce bHaBac-Δvfgf-egfp-ph. The ph gene together with vfgf under the control of its own promoter were inserted at the ph locus of bHaBac-Δvfgf-egfp to generate bHaBac-REPvfgf-egfp-ph. Previously constructed bHaBac-egfp-ph containing the egfp-cat cassette and ph gene was used as a positive control. (B) Transfection-infection assays of recombinant bacmids. Each recombinant bacmid DNA was transfected into HzAM1 cells (a, b, and c). At 6 d.p.t., clarified supernatant was used to infect healthy HzAM1 cells (d, e and f). The transfected and infected cells were monitored by fluorescence microscopy. (C) Western blot detection of recombinant viruses. BVs of each recombinant virus were collected from the supernatant of infected cells, purified by ultracentrifugation, and electrophoresed by 10% SDS-PAGE. The expression of vFGF was detected with anti-vFGF antibody, and the major capsid protein VP39 was used as an internal control. M, protein molecular marker; d.p.i., days post-infection.
-
To construct the vfgf-repaired bacmid, the vfgf gene under its native promoter was PCR amplified with the primers vfgf-f and vfgf-r (Supplementary Table S1) from bHaBacHZ8 using Pyrobest DNA Polymerase (TaKaRa, Kusatsu, Japan). The PCR product was cloned into the pGEM-T Easy vector (Promega) and subjected to sequence confirmation. The vfgf fragment was digested from the vector and cloned into the transfer vector pFB-DUAL-ph (Song et al., 2008) to generate pFB-DUAL-vfgf-ph. The expression cassette was transposed into bHaBac-Δvfgf-egfp by Tn7-mediated transposition according to the Bac-to-Bac Baculovirus Expression Systems manual (Invitrogen), and the resulting bacmid was designated as bHaBac-REPvfgf-egfp-ph (Figure 2A). The polyhedrin (ph) gene was reintroduced into bHaBac-Δvfgf-egfp to produce bHaBac-Δvfgf-egfp-ph using the previously constructed pFB-DUAL-ph, since the original ph was disrupted during the construction of bHaBacHZ8 (Figure 2A). The previously constructed bHaBac-egfp-ph containing the egfp-cat cassette and ph was used as a positive control (Song et al., 2008).
-
HzAM1 cells (5.0×105/well) were transfected with 2 μg DNA of each recombinant bacmid using 12 μL of Lipofectin reagent (Invitrogen) according to the Bac-to-Bac Baculovirus Expression Systems manual. At 6 days post-transfection (d.p.t.), cell supernatants were harvested by centrifugation at 945×g for 5 min and then used to infect another batch of HzAM1 cells. Green cells were monitored by fluorescence microscopy daily for the successful transfection and spread of infection. For BV amplification, HzAM1 cells were infected at an MOI of 0.1, and the supernatant was collected at 6 days post-infection.
-
For the virus growth curves, 3×105 HzAM1 cells were infected with vHaBac-Δvfgf-egfp-ph, vHaBac-REPvfgf-egfp-ph, or vHaBac-egfp-ph at an MOI of 10. Supernatants were harvested at 0, 12, 24, 48, 72, and 96 h.p.i., and BV titers were determined by end-point dilution assays (EPDAs). Infection experiments were performed in triplicate, and the mean BV titers were analyzed using one-way analysis of variance (SPSS Inc., Chicago, IL, USA) with virus type and time as factors (Yin et al., 2008).
-
To detect viral DNA replication in infected cells, HzAM1 cells (3×105) were infected with vHaBac-Δvfgf-egfp-ph or vHaBac-egfp-ph at an MOI of 10. Total cellular DNA was isolated at 0, 24, and 48 h.p.i. using Genomic DNA Rapid Isolation Kit (BioDev, Beijing, China). Primers Ha39-F and Ha39-R designed previously were used to determine viral copy numbers in total cellular DNA according to previously reported methods (Huang et al., 2014).
-
To investigate the effect of vfgf deletion on HearNPV infectivity, bioassays were performed as described previously (Sun et al., 2009; Luo et al., 2011). The median lethal dose (LD50) of ODV was determined with third-instar H. armigera larvae using the food-contamination method with eight different doses of occlusion bodies (OBs): 1×103, 3×103, 1×104, 3×104, 1×105, 3×105, 1×106, and 3×106. After inoculation, the larvae were transferred to a fresh diet and maintained separately at 27℃ in 24-well plates. Twenty-four insects were inoculated for each dose, and mortality was checked daily. Each test was performed twice. The data from two replicates were pooled to calculate the final LD50 values when there was no significant difference between replicate results. LD50 values and the confidence limits of different variants were determined by probit analysis (SPSS Inc.) and compared by standard lethal-dose ratio comparison (Robertson et al., 2007).
The median survival time (ST50) of third-instar H. armigera larvae dosed with different variants was determined using a food-contamination method as described previously (Luo et al., 2011). Briefly, 1×106 OBs were applied to a small piece of artificial diet and after completely ingesting the contaminated food, the larvae were further reared using an uncontaminated diet. Forty-eight insects were tested for each virus, and mortality was checked every 6 h. ST50 values were calculated using a Kaplan-Meier estimator and compared using the log-rank test.
Virus, cell line, and insects
Antibody, transcription, and expression analysis
Construction of the vfgf-null HearNPV bacmid
Construction of the vfgf-repaired bacmid
Transfection and infection assay
One-step growth-curve analysis
Viral DNA-replication assay
Bioassays
-
The fgf gene of HearNPV encodes a ~40-kDa protein containing 302 amino acids and six predicted N-linked glycans (Figure 1A). An early-transcription-initiation motif, TATATAA, followed by CAGT (18 nt downstream from TATA), was found 54 nt upstream of the ATG translation start codon of vfgf (Figure 1A), suggesting that vfgf is an early gene. In fact, we detected vfgf transcripts (575 bp) by reverse transcription (RT)-PCR analysis at 3 h.p.i. (Figure 1B), and western blot analysis indicated a protein band with the expected size of vFGF (~40 kDa) at 16 h.p.i. (Figure 1C). Given that HearNPV genomic DNA replication begins at ~14 h.p.i. (Dai et al., 2006), vfgf was suggested to be an early gene of HearNPV, with its expression continuing into the later stages of infection.
-
To determine vfgf function in HearNPV infectivity, the vfgf gene from bHaBacHZ8 was inactivated by homologous recombination and replaced by an egfp-cat cassette, with the resulting bacmid designated as bHaBac-Δvfgf-egfp (Figure 2A). Two recombinant bacmids were constructed based on bHaBac-Δvfgf-egfp: a vfgf-knockout bacmid (bHaBac-Δvfgf-egfp-ph), where the ph gene was re-inserted into the ph locus; and a vfgf-repaired bacmid (bHaBac-REPvfgf-egfp-ph), where the ph gene and vfgf under the control of its own promoter were inserted into the ph locus (Figure 2A). A previously constructed bacmid (bHaBac-egfp-ph) containing egfp and ph was used as a positive control (Luo et al., 2011). Using a transfection-infection assay, the effect of vfgf deletion on BV propagation was monitored by fluorescence microscopy (Figure 2B). Infection was observed in cells incubated with bHaBac-Δvfgf-egfp-ph-transfected cell supernatant, indicating that vfgf is nonessential for in vitro viral replication and infection (Figure 2B-a and B-d). Transfection-infection of the vfgf-repaired bacmid and the positive-control bacmid revealed results similar to those observed with bHaBac-REPvfgf-egfp-ph (Figure 2B-b, c, e and f).
The absence of vFGF in HaBac-Δvfgf-egfp-ph BVs was confirmed by western blot analysis. With anti-vFGF antiserum, no specific band was detected in BV samples harvested from the supernatant of vHaBac-Δvfgf-egfp-ph-infected HzAM1 cells. In contrast, a band of ~40 kDa was clearly detected in BV samples from vHaBac-REPvfgf-egfp-ph-and vHaBac-egfp-ph-infected cells (Figure 2C). The expression of VP39 indicated that similar BV concentrations were loaded into each lane (Figure 2C). These results confirmed the correct construction of all of the recombinants.
-
To study the effect of vfgf on infectious BV production, one-step growth curves were determined and compared. HzAM1 cells were infected with vHaBac-Δvfgf-egfp-ph, vHaBac-REPvfgf-egfp-ph, or vHaBac-egfp-ph in parallel (MOI=10). Supernatants collected at different post-infection time points were titrated by EPDA. The results showed that vHaBac-Δvfgf-egfp-ph, vHaBac-REPvfgf-egfp-ph, and vHaBac-egfp-ph exhibited similar kinetics of infectious BV production (Figure 3A). Statistical analysis also revealed that there was no significant difference between vfgf-knockout and -repaired viruses or with the control virus at all infection time points (P > 0.05).
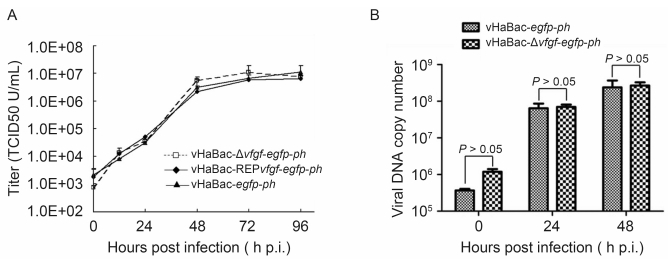
Figure 3. Effect of vFGF on viral growth kinetics and DNA replication. (A) One-step growth-curve analysis of BV production. HzAM1 cells were infected with either vHaBac-Δvfgf-egfp-ph, vHaBac-REPvfgf-egfp-ph, or vHaBac-egfp-ph at an MOI of 10. BVs were harvested at the indicated post-infection time points and titrated onto HzAM1 cells. Each data point represents the average titer from three independent infections, and error bars represent standard deviations. (B) Real-time PCR analysis of viral DNA replication. HzAM1 cells were infected with the respective virus at an MOI of 10. At the indicated post-infection time points, total cellular DNA was isolated, digested with Dpn I, and subjected to real-time PCR analysis. The experiments were performed in triplicate, and error bars represent standard deviations. TCID50, 50% tissue-culture infection dose.
To further illustrate the effect of vfgf deletion on viral DNA synthesis, the level of viral DNA replication during infection was monitored by real-time PCR analysis. The results indicated that there was no significant difference in viral DNA copy number between vHaBac-Δvfgf-egfp-ph-and vHaBac-egfp-ph-infected cells at both 24 and 48 h.p.i. (P > 0.05; Figure 3B). These results indicated that vfgf deletion had no significant influence on either infectious BV production or viral DNA replication in cultured cells.
-
To determine the LD50 value of vHaBac-Δvfgf-egfp-ph, vHaBac-REPvfgf-egfp-ph, and vHaBac-egfp-ph, third-instar H. armigera larvae were infected orally with various doses of OBs and monitored for mortality. The LD50 value of vHaBac-Δvfgf-egfp-ph was 16.9 (12.4, 22.7) ×103 OBs, which was significantly higher than that of vHaBac-REPvfgf-egfp-ph or vHaBac-egfp-ph (P < 0.05), while no significant difference was observed between the LD50 values of vHaBac-REPvfgf-egfp-ph or vHaBac-egfp-ph (P > 0.05) (Table 1). We also compared the time required to kill infected larvae, with the results indicating that the ST50 value of vHaBac-Δvfgf-egfp-ph (~107 h) was significantly higher than that of vHaBac-egfp-ph (~95 h; P < 0.05), while no significant difference was observed between the ST50 values of vHaBac-REPvfgf-egfp-ph or vHaBac-egfp-ph (P > 0.05). These data indicated that vfgf deletion resulted in a delayed time of death of larvae infected orally (Table 2). Taken together, these results suggested that vfgf deletion significantly reduced HearNPV infectivity in larvae.
Viruses LD50×103 OBs (95% CI) Potency ratioa (95% CI) vHaBac-egfp-ph 9.2 (6.7, 12.5) - vHaBac-Δvfgf-egfp-ph 16.9 (12.4, 22.7) 1.8* (1.2, 2.9) vHaBac-REPvfgf-egfp-ph 8.7 (6.4, 12.0) 0.9 (0.6, 1.5) Note: a Potency ratio was calculated by dividing the LD50 of the vfgf-deleted or -repaired variants by that of vHaBac-egfp-ph. * Indicates significant difference based on the 95% CI of the potency ratio, including the value 1.0 (Robertson et al., 2007). CI, confidence interval. Table 1. Dose-mortality responses of vHaBac-egfp-ph, vHaBac-Δvfgf-egfp-ph, and vHaBac-REPvfgf-egfp-ph in third-instar H. armigera larvae
Tests Viruses ST50 (95% CI) (h) χ2 P 1 vHaBac-egfp-ph 95 (93.2, 96.7) - - vHaBac-Δvfgf-egfp-ph 107.5 (103.1, 111.9) 27.834 1×10-6 * vHaBac-REPvfgf-egfp-ph 94.5 (93.1, 95.9) 3.568 0.059 2 vHaBac-egfp-ph 95 (93.1, 96.9) - - vHaBac-Δvfgf-egfp-ph 107.5 (104.5, 110.5) 19.915 8×10-6* vHaBac-REPvfgf-egfp-ph 94.5 (93.6, 95.4) 3.327 0.068 Note: * Indicates significant difference between the vfgf-deleted variant and vHaBac-egfp-ph based on log-rank test. CI, confidence interval. Table 2. Time-mortality responses of vHaBac-egfp-ph, vHaBac-Δvfgf-egfp-ph, and vHaBac-REPvfgf-egfp-ph in third-instar H. armigera larvae.
Transcription and expression analysis of HearNPV vFGF
Construction and verification of vfgf-deleted and -repaired HearNPVs
The effect of vfgf deletion on infectious BV production and viral DNA replication in vitro
The contribution of vFGF to viral infectivity in infected insects
-
The vfgf gene is conserved in most lepidopteran baculoviruses sequenced to date. The function of the vfgf of Group Ⅰ alphabaculovirus has been studied and was suggested to play a role in dissemination of viral infection from the midgut of the insect host (Detvisitsakun et al., 2006; Katsuma et al., 2006b; Detvisitsakun et al., 2007). The function of vFGF was also suggested to be dependent upon activation of matrix metalloproteases and effector caspases, leading to BL alteration and stimulation of tracheoblast and midgut epithelial cell migration (Means and Passarelli, 2010). Compared to Group Ⅰ alphabaculovirus vFGF, the vFGF of Group Ⅱ alphabaculoviruses has a larger C-terminal region and more putative N-glycosylation sequences. Here, we investigated the function of HearNPV vFGF, with results indicating that vfgf deletion had no clear impact on BV production or viral DNA replication in cultured cells. In the host insects, the lack of vfgf resulted in reduced infectivity and a delayed time of death of larvae when delivered by feeding.
The transcription and expression pattern of HearNPV vfgf was investigated at various time points following HearNPV infection. The transcription of vfgf was detected as early as 3 h.p.i., suggesting that vfgf is an HearNPV early gene (Figure 1B), and a baculovirus consensus early promoter was also found in the upstream region of the vfgf-translation start codon. Additionally, BmNPV vfgf was identified as an early gene, although no typical baculovirus promoter motifs were found (Katsuma et al., 2004). For some viruses, such as Epiphyas postvittana MNPV and Chrysodeixis chalcites NPV, no consensus baculovirus vfgf-promoter motifs were detected (Hyink et al., 2002; Oers et al., 2005), while baculovirus late-promoter motifs were found associated with the vfgf from Ectropis obliqua nucleopolyhedrovirus, Mamestra configurata NPV, and Lymantria dispar multinucleocapsid NPV (Kuzio et al., 1999; Li et al., 2002; Ma et al., 2007). This suggested that vfgf transcriptional regulation may vary among baculoviruses, and that the transcription pattern may not be strictly related to vfgf phylogeny.
Genomic sequence comparison revealed that most alphabaculoviruses encode one vfgf homolog, even though there are three vfgf homologs (vFGF-1, 2, and 3) identified in betabaculovirus. (Yin et al., 2015). Sequence analysis revealed that HearNPV vFGF contains a typical FGF-superfamily central motif of ~120 amino acids essential for binding to an FGF receptor, which is also present in Group Ⅰ alphabaculovirus vFGFs (Figure 1A) (Popovici et al., 2005). In contrast, the FGF-superfamily central motif of betabaculovirus vFGFs is much shorter (~30 amino acids) and nontypical (data not shown). Therefore, HearNPV vFGF appears to share greater similarity with Group Ⅰ alphabaculovirus vFGFs relative to those from betabaculoviruses based on conservation of a structural domain and potential function. Functional analysis indicated that vfgf deletion did not influence ODV morphogenesis or occlusion (data not shown), but significantly impaired HearNPV infectivity in larvae (Table 1 and 2). We speculated that HearNPV vFGF may also play a role in dissemination of virus infection from the midgut as proposed in previous studies of Group Ⅰ vFGFs; however, confirmation requires further evidence.
Although AcMNPV and BmNPV are closely related Group Ⅰ alphabaculoviruses, the function of their vFGFs differ significantly. A study of AcMNPV revealed that a vfgf knockout exhibited no significant in vitro effect on DNA and protein syntheses or BV production (Detvisitsakun et al., 2007). In contrast, vfgf deletion in BmNPV reduced BV production in both cultured cells and infected larvae (Katsuma et al., 2006b). Furthermore, the expression of late and very late genes was also reduced in vfgf-deficient BmNPV (Katsuma et al., 2006b). In our study, we observed that HearNPV vfgf deletion did not influence either BV production (Figure 3A) or the expression of late and very late viral genes (data not shown), implying that the in vitro function of HearNPV vFGF is much closer to that reported for AcMNPV as compared to BmNPV. In larval bioassays, both the LD50 and ST50 values of vfgf-deleted HearNPV increased; however, only the latter increased in vfgf-deleted AcMNPV or BmNPV. The different phenotypes associated with vFGF-deleted baculoviruses may be determined by the coaction of different vFGFs and host species. We are currently constructing recombinant HearNPVs wherein the vfgf gene is replaced with counterparts from other viruses in order to more effectively compare the functions of vFGFs in the same viral backbone and host systems.
In summary, this is the first report concerning the function of vFGF from a Group Ⅱ alphabaculovirus. Our results indicated that HearNPV vFGF transcription begins at the early stage of infection and continues into the late stage of infection. These in vitro experiments showed that the lack of vfgf did not influence HearNPV viral growth kinetics or DNA replication, while vfgf deletion significantly impaired HearNPV infectivity in larvae. Therefore, vFGF represents an important virulent factor associated with in vivo HearNPV infection.
-
This work was supported by grants from the National Science Foundation of China (No. 31200124 and 31321001) and the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB11030400). We acknowledge the Core Facility and Technical Support of Wuhan Institute of Virology for technical assistance. The authors would like to thank Dr. Xiulian Sun for the great help in statistical analysis.
-
The authors declare that they have no conflicts of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
-
FFY, ZHH, HLW, and MLW designed the experiments. FFY, RKD, WHK, and GY performed the experiments. FFY, FD, and HLW analyzed the data. FFY, ZHH, and MLW wrote the paper. All the authors approved the final manuscript.
Supplementary Table are available on the website of Virologica Sinica: www.virosin.org; link.springer.com/journal/12250.
-
Primer Primer sequence (5′-3′) anti-vFGF-f GCGGGATCCAGACCGGGCGGACGAAAC anti-vFGF-r GCGAAGCTTCATGTACGTTACAACAACAG vFGF trans-f CGTGCCGTTATATTTGCGTTAACCAATGC vFGF trans-r CAGAGGTAACATGGATGATGTCTGTGG del-F1f CGGGGTACCATACAAATTCAACCTGCGAACTC del-F1r CCGCTCGAGATCATCCATGTTACCTCTGAC del-F2f CCGGATATCGTGCGAGTCAGTAGAATTTGTT del-F2r CTAGTCTAGAATTTCTAACACATTCTATGCGATTG vfgf-f GCGCTCGAGTGGAGCAGATTCAAGATTCGCC vfgf-r GCGGGTACCCATGTACGTTACAACAACAG Table S1. Sequences of primers used for PCR amplification














 DownLoad:
DownLoad: