-
Human cytomegalovirus (HCMV) has abilities to infect a broad range of cell types in human being. After primary infection, HCMV can maintain a persistent or latent infection in its host lasting a whole lifetime. HCMV infection in immunocompromised individuals, such as transplant recipients or AIDS patients, can also lead to multiple serious complications (Einsele and Hebart, 1999; Khoshnevis and Tyring, 2002).
MicroRNAs (miRNAs) are a class of small noncoding RNA molecules that play a role in regulating gene expression by controlling target mRNA translation or degradation (Zhao and Srivastava, 2007). As important post-transcriptional regulators, miRNAs are involved in the regulation of diverse cellular processes, such as cell development, cell differentiation, proliferation, metabolism, cell cycle and apoptosis (Klooster et al., 2006). To date, at least 26 mature miRNAs were identified during HCMV lytic infection (Babu et al., 2014). Recent studies demonstrated that some of the HCMV encoded miRNAs, including hcmv-miR-US5-1, are also expressed during latency (Shen et al., 2014; Fu et al., 2014). Hcmv-miR-US5-1 was reported to regulate HCMV gene US7 synergistically with hcmv-miR-US5-2 (Tirabassl et al., 2011). Unfortunately, the unknown functions of HCMV US7 gene hinder the determination of the exact effects of hcmv-miR-US5-1 on HCMV or its host cells. Recently, hcmv-miR-US5-1 was reported to target secretary pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion synergistically with hcmv-miR-UL112-1 and hcmv-miR-US5-2 (Hook et al., 2014). The expression of hcmv-miR-US5-1 during lytic and latent infections suggests that it may play an important role during HCMV infection. Identifying the target genes and regulatory functions of hcmv-miR-US5-1 may provide further insight into the study of HCMV pathogenesis.
In our study, the DNA replication inhibitor, GMNN, which accumulates during HCMV infection and plays a critical role in cell cycle regulation (Wu et al., 2014; Tada et al., 2007; Ballabenu et al., 2013; Biswas et al., 2003), was identified to be a direct target of hcmv-miR-US5-1. Over-expression of hcmv-miR-US5-1 was demonstrated to reduce HCMV DNA copies in infected cells by blocking the accumulation of GMNN. At the same time, it was found that consequent lower expression of GMNN could modulate host cell cycle and promote host cell proliferation.
-
Human embryonic lung fibroblasts cells (MRC-5) and human brain glioma cells (U373) used in our study were acquired from Shanghai Institute for Biological Sciences, Chinese Academy of Sciences (CAS). Human embryonic kidney cell line HEK293 was a gift from Department of Medical Genetics, China Medical University. MRC-5 cells were maintained in Modified Eagle’s Medium (MEM) supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin and streptomycin. U373 and HEK293 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FBS, 2 mmol/L L-glutamine, 100 units/mL penicillin and streptomycin. All cell cultures were kept at 37 °C in a 5% CO 2 incubator.
-
Han was a low-passaged HCMV clinical strain, which was isolated from a urine sample of a 5-month-old infant hospitalized in Shengjing Hospital of China Medical University. The collection was undertaken with the understanding and written consent of the subject’s parents, and was approved by the ethical committee of Shengjing Hospital. Han strian was propagated in human embryonic lung fibroblasts cells maintained in MEM supplemented with 2% FBS and penicillin-streptomycin. The supernatant of the infected cells was collected, centrifuged and stored at –80 °C till use.
-
MRC-5 cells were plated in 60 mm plates and then were infected with Han stock at 3 multiplicity of infection (MOI). Total cellular RNAs were isolated using TRIzol (Invitrogen, Waltham, USA) at 0, 24, 48, 72, 96, 120 and 144 hours post infection (hpi) respectively, and then treated with TURBO DNA-freeTM Kit (Ambion, Waltham, USA). RNAs were reverse transcript with hcmv-miR-US5-1 specific primer (Applied Biosystems, Waltham, USA) using TaqMan miRNA reverse transcription kit (Applied Biosystems). Quantative PCR was performed using the Universal PCR Master Mix Kit and hcmv-miR-US5-1 specific probe (Applied Biosystems) on 7300 Fast Real-time PCR System. snRNA U6 was also amplified as a normalization control. The relative expression level was calculated using 2–ΔΔCT method (Schmittgen et al., 2008).
-
Hcmv-miR-US5-1 putative targets were screened from HCMV Han infected MRC5 cells using hybrid-PCR as previously described (Huang et al., 2011). The hybrid-PCR primer for hcmv-miR-US5-1 (5′-RCGCTCTCGTCRGGCTTGTC-3′) was designed according to its sequence. The letter R in primer sequence represents ether adenine (A) or guanine (G). The products from hybrid-PCR were purified using Promega Wizard SV Gel and PCR Clean-up System (Promega, Waltham, USA), and were then ligated into pMD-19T vectors (TakaRa, Waltham, USA). One hundred and sixteen clones were randomly selected and the inserts were sequenced on an ABI 3730 automated sequencer. The mRNA specific sequences located between the hcmv-miR-US5-1 hybrid-primer binding sites and polyA structures were intercepted to identify the putative target genes online (http://www.ncbi.nlm.nih.gov/blast).
-
Eleven putative target mRNAs were chosen for further validation by luciferase reporter assays. Sequences of these target mRNAs which containing hcmv-miR-US5-1 putative binding sites were amplified directly from mRNA-derived cDNA. The products were then cloned into luciferase reporter vector pMIR between the Spe I and Hind III sites (Ambion, Waltham, USA), respectively. The mutant plasmid pMIR-GMNN-Mut, which contains substitution nucleotides from GCTT to TGTA at corresponding binding site of hcmv-miR-US5-1 seed region, was constructed by a Site-directed Gene Mutagenesis Kit (Beyotime, Shanghai, China) based on the wide type plasmid pMIR-GMNN-Wt. Primer sequences used in plasmid construction were listed in Supplementary Table S1. All constructs were confirmed by DNA sequencing.
-
HEK293 cells were prepared in 24-well plates and reached a confluence of 85% at the time of transfection. 0.2 μg plasmids (empty vector pMIR, pMIR-GMNN-Wt or the mutant plasmid pMIR-GMNN-Mut) were transfected respectively into the HEK293 cells along with 100 nmol/L hcmv-miR-US5-1 mimics (RiboBio, Guangzhou, China) and 0.2 μg control renilla plasmid pRL-TK (Promega, Wisconsin, USA) using Lipofectamine 2000 (Lipo2000) (Invitrogen). A mimic which produces a random small RNA was used as a negative control. Luciferase activities in each well were measured using Dual Luciferase Reporter Assay System (Promega) 48 hours after transfection according to the manufacturer’s instructions. All measurements were done in triplicate wells and signals were normalized against the renilla control. Data from three independent repetitions were presented as the mean ± SD and used in statistical analyses.
-
Proteins were extracted in our study using Mammalian Protein Extraction Reagent M-PER (ThermoScientific, Waltham, USA) according to the manufacturer’s instructions. Before electrophoresis, proteins were quantified using a BCA protein assay kit (Beyotime). The extracted proteins were separated in a 10% acrylamide gel and transferred onto polyvinylidene fluoride (PVDF) membranes. Western blot analyses were performed using GMNN or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) specific antibodies (Abcam, Cambrideg, USA). Immunoblots were visualized via an electrochemiluminescence (ECL) detection system.
-
U373 cells were plated in 6-well plates and transfected respectively with 100 nmol/L miRNA negative control, 100 nmol/L hcmv-miR-US5-1 mimics (RiboBio, Guangzhou, China), 100 nmol/L hcmv-miR-US5-1 specific inhibitor (RiboBio, Guangzhou, China) and 40 nmol/L GMNN siRNA using Lipo2000. The transfected U373 cells were inoculated with Han strain at 3 MOI 24 hours post transfection. Total cellular DNA was extracted at 0, 24, 48, 72 hpi. HCMV DNA copies were then determined by quantative PCR assay using a commercial kit (Da An Gene, Guangzhou, China). All measurements were done in triplicate and the results from three independent repetitions were presented as mean ± SD. Meanwhile, expressions of endogenous GMNN and hcmv-miR-US5-1 in the U373 cells were detected as described above.
-
U373 cells in 6-well plates were synchronized in low-serum medium (0.2%) for 48 hours. And then the cells were transfected respectively with 100 nmol/L miRNA negative control, 100 nmol/L hcmv-miR-US5-1 mimics, a mixture of 100 nmol/L hcmv-miR-US5-1 mimics and 300 nmol/L hcmv-miR-US5-1 specific inhibitor, and 40 nmol/L GMNN siRNA using Lipo2000. The cells were collected by trypsinization 48 hours after transfection. Cell cycles were analyzed using a Cell Cycle and Apoptosis Analysis Kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions on a FACScan Flow cytometer (Beckton Dickinson). The data from 20, 000 cells was collected for analysis.
-
After synchronization, U373 cells in 96-well plates were transfected respectively with 100 nmol/L miRNA negative control, 100 nmol/L hcmv-miR-US5-1 mimics, a mixture of 100 nmol/L hcmv-miR-US5-1 mimics and 300 nmol/L hcmv-miR-US5-1 specific inhibitor, and 40 nmol/L GMNN siRNA using Lipo2000. Cell proliferation was assessed at 48 hours after transfection using a Cell Counting Kit (Fanbo Biochemicals, Beijing, China) according to the manufacture’s instructions. The absorbance was measured at 450 nm using a 96 well plate reader (Dynex technologies, Chantilly, USA). Measurements were done in triplicate and the results from three independent repetitions were presented as mean ± SD.
-
Data are shown as mean ± SD. Statistical significance was determined by Student’s t-test, with P-value of < 0.05 considered to be statistically significant.
-
In this study, expression kinetics of hcmv-miR-US5-1-5p was determined in U373 cells at different time points during HCMV infection. As shown in Figure 1, hcmv-miR-US5-1 was highly expressed 24 hpi and then accumulated gradually along with prolonged HCMV infection.
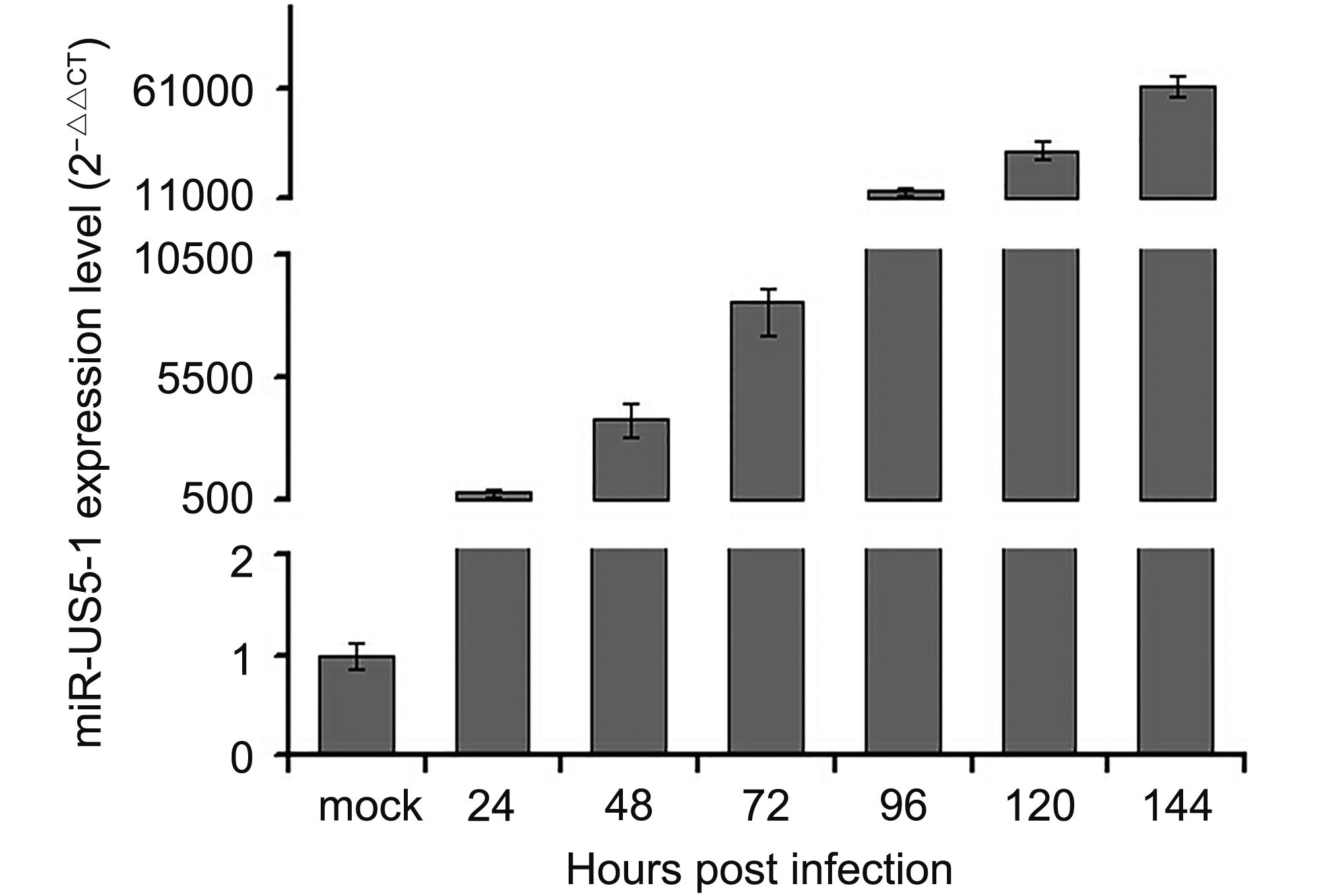
Figure 1. Expression kinetics of hcmv-miR-US5-1 in HCMV infected U373 cells. U373 cells were infected with HCMV at 3 MOI. Expression levels of hcmv-miR-US5-1 in HCMV infected cells were quantified by RT-qPCR at 0, 24, 48, 72, 96, 120 and 144 hpi. Data from three independent repetitions were used for statistical analysis.
-
Hybrid-PCR was used to identify putative targets of hcmv-miR-US5-1. Specific sequences of 23 candidate target mRNAs were obtained from 116 sequenced clones (Table 1). Corresponding mRNAs were identified by blast in GenBank database (http://www.ncbi.nlm.nih.gov/GenBank). All identified putative target mRNAs of hcmv-miR-US5-1 were located in host genome. We then validated a number of mRNAs by luciferase reporter assay. Eleven putative target mRNAs were chosen from our results above, including three mRNAs whose target sites located in coding regions. As shown in Figure 2, led to a diversity decrease of luciferase activity was observed in samples transfected with hcmv-miR-US5-1 and pMIR vectors containing candidate target sequences compared to the miRNA negative control group (NC). These results demonstrate that hcmv-miR-US5-1 can directly bind to these putative target sequences that obtained from hybrid-PCR results. Notably, the putative target mRNAs NUDT5, GMNN and RPL5 whose target sites located in CDS were also recognized and combined by hcmv-miR-US5-1. Among these target genes, GMNN, the cellular DNA replication inhibitor was previously reported to accumulate during HCMV infection. We further investigate the regulation between hcmv-miR-US5-1 and GMNN.
Putative target mRNA Accession Number Position of binding site Nudix (nucleoside diphosphate linked moiety X)-type motif 5 (NUDT5) NM_014142.2 CDS Geminin, DNA replication inhibitor (GMNN) NM_001251989.1 CDS Ribosomal protein L5 (RPL5) NM_000969.3 CDS Polycomb group ring finger 2 (PCGF2) NM_007144.2 3′ UTR Serine/threonine/tyrosine interacting-like 1 (STYXL1) NM_016086.2 3′ UTR Deleted in liver cancer 1 (DLC1) NM_182643.2 3′ UTR SIN3 transcription regulator family member B (SIN3B) NM_015260.2 3′ UTR Ribosomal protein S18 (RPS18) NM_022551.2 3′ UTR Ornithine decarboxylase antizyme 1 (OAZ1) NM_004152.2 3′ UTR Nuclear factor I/X (CCAAT-binding transcription factor) (NFIX) NM_002501.3 3′ UTR Apoptotic peptidase activating factor 1 (APAF1) NM_013229.2 5′ UTR ATPase, H+ transporting, lysosomal 14kDa, V1 subunit F (ATP6V1F) NM_001198909.1 3′ UTR Transmembrane protein 47(TMEM47) NM_031442.3 3′ UTR Transmembrane protein 109 (TMEM109) NM_024092.2 3′ UTR Inner membrane protein, mitochondrial (IMMT) NM_006839.2 3′ UTR Ferritin, heavy polypeptide 1 (FTH1) NM_002032.2 CDS Tripartite motif containing 26 (TRIM26) NM_003449.4 3′ UTR Ral guanine nucleotide dissociation stimulator-like 1 (RGL1) NM_015149.3 3′ UTR Zinc finger, SWIM-type containing 7 (ZSWIM7) NM_001042697.1 3′ UTR Ribosomal protein L41 (RPL41) NM_021104.1 3′ UTR RAD51 paralog D (RAD51D) NM_002878.3 3′ UTR KIAA1462 NM_020848.2 3′ UTR Chromosome 20 open reading frame 194 (C20orf194) NM_001009984.2 3′ UTR Table 1. Putative targets of hcmv-miR-US5-1 identified by hybrid-PCR
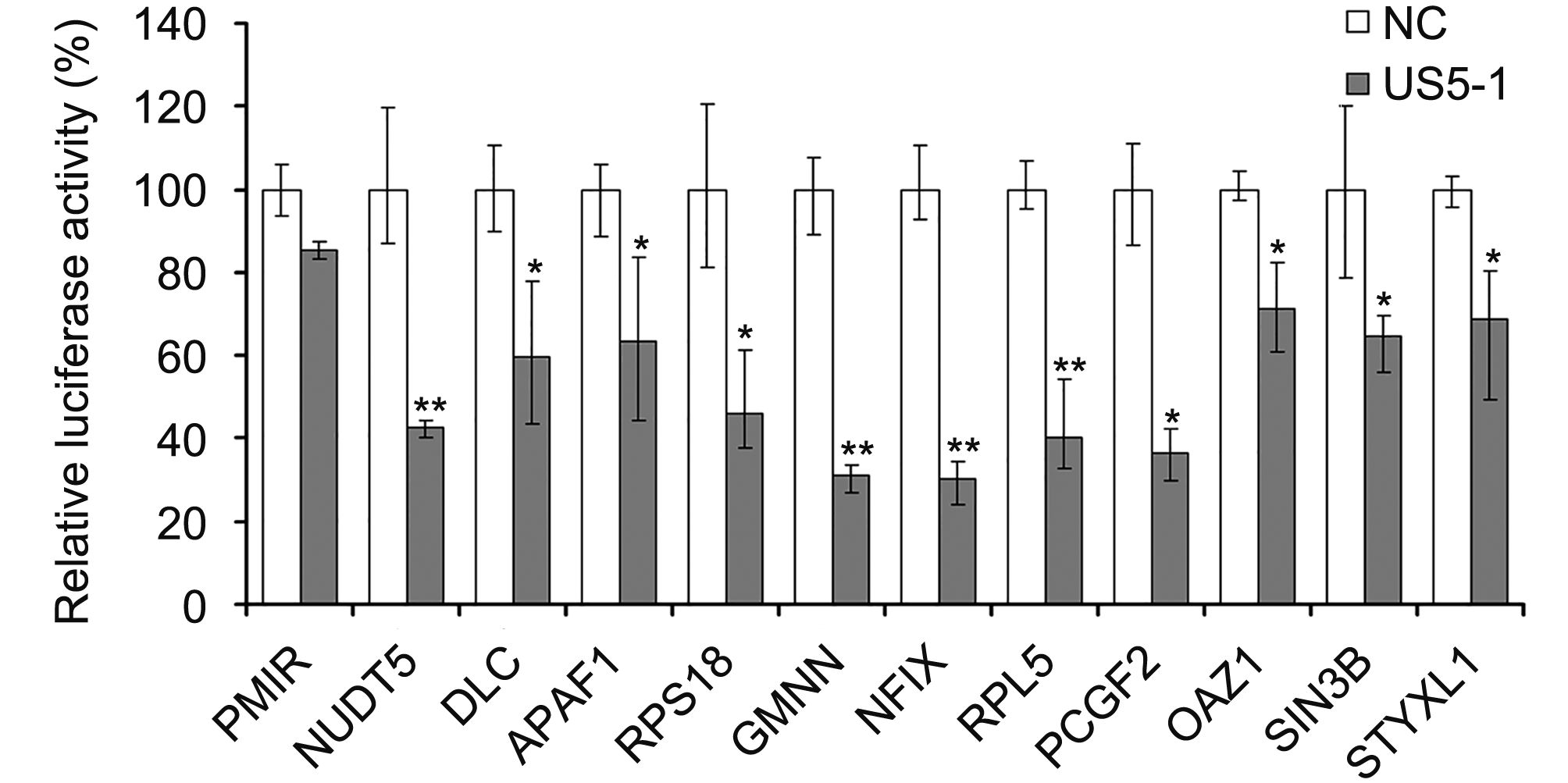
Figure 2. Hcmv-miR-US5-1 mediated repression of target luciferase activity. Dual luciferase assays in HEK293 cells transfected respectively with the empty PMIR reporter vector (pMIR), the reporter vectors containing the putative target sequences together with miRNA negative control (NC) or hcmv-miR-US5-1 (US5-1). Results were shown as percentage expression of negative control sample (NC) following correction for transfection levels according to control renilla luciferase expression. Data from three independent repetitions were used for statistical analysis. ** indicates P < 0.01, * indicates P < 0.05, by comparing to data from cells transfected with miRNA negative control.
-
The predicted binding site for hcmv-miR-US5-1 was located in nucleotides (nt) 128–148 in GMNN coding region. This binding site was also predicted by online software RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html) with minimum free energy (mfe) of -29.8kcal/moL (Figure 3A). To further validate whether GMNN is a direct target of hcmv-miR-US5-1, luciferase reporter assay and western blot were performed. As shown in Figure 3B, compared those negative control, cells transfected with pMIR-GMNN-Wt and hcmv-miR-US5-1 mimics resulted in a significant decrease (~75%) of luciferase activity; however, mutation in the hcmv-miR-US5-1 binding site could abolish the effect of hcmv-miR-US5-1 (Figure 3B). Ectopic expression of hcmv-miR-US5-1 markedly reduced the endogenous GMNN protein levels in U373 cells at the same level of those treated with specific siRNA for GMNN. Silencing the over expressed hcmv-miR-US5-1 by its specific inhibitor counteracted the inhibition effect of hcmv-miR-US5-1 on GMNN expression (Figure 3C). These data demonstrated that hcmv-miR-US5-1 could specifically inhibit GMNN protein expression through binding to the coding sequence of GMNN.
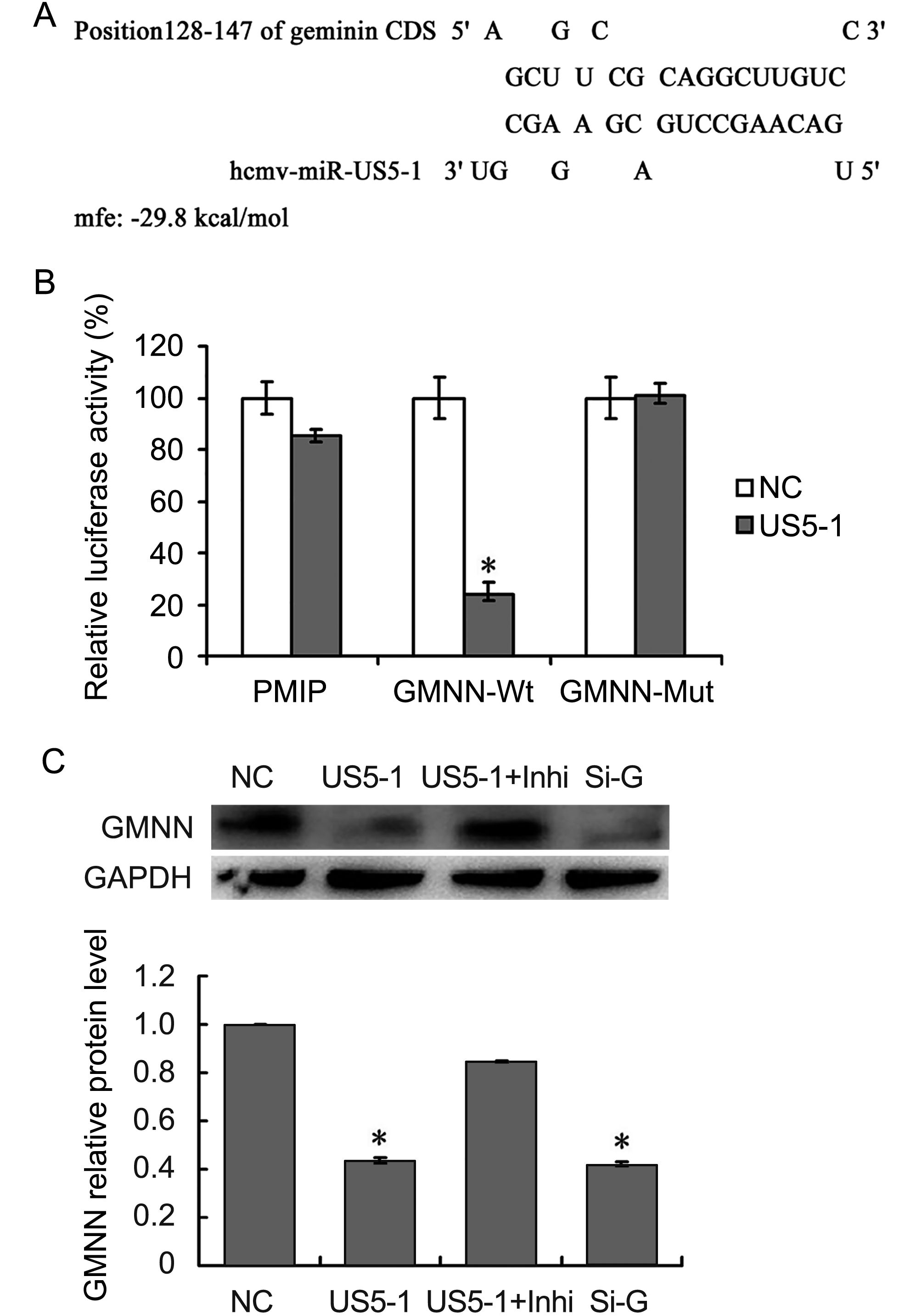
Figure 3. Hcmv-miR-US5-1 mediated inhibition of GMNN expression by targeting GMNN coding sequence. Schematic diagram of predicted binding site of hcmv-miR-US5-1 in the GMNN coding domain sequence (CDS). The binding site is at nucleotide 128–148 of the GMNN CDS. (B) Dual luciferase assay were performed in HEK293 cells transfected respectively with the empty PMIR reporter vector (PMIR), the reporter vectors containing the wild type GMNN CDS (GMNN-Wt) and mutant type GMNN CDS (GMNN-Mut) together with miRNA negative control (NC) or hcmv-miR-US5-1 (US5-1). (C) Expressions of GMNN in U373 cells transfected respectively with miRNA negative control (NC), hcmv-miR-US5-1 mimics (US5-1), hcmv-miR-US5-1 mimics and its inhibitor (US5-1+Inhi), and siRNA for GMNN (Si-G) were detected by western blot. Data from three independent repetitions were used for statistical analysis.* indicates P < 0.05, by comparing to data from cells transfected with miRNA negative control.
-
GMNN proteins were detected in U373 cells mock or infected with HCMV in different conditions. As shown in Figure 4, the expression of GMNN was increased in HCMV infected cells compared with mock cells, and its level accumulated as the infection progressed. However, after transfection of hcmv-miR-US5-1 mimics, the accumulation of GMNN was blocked.
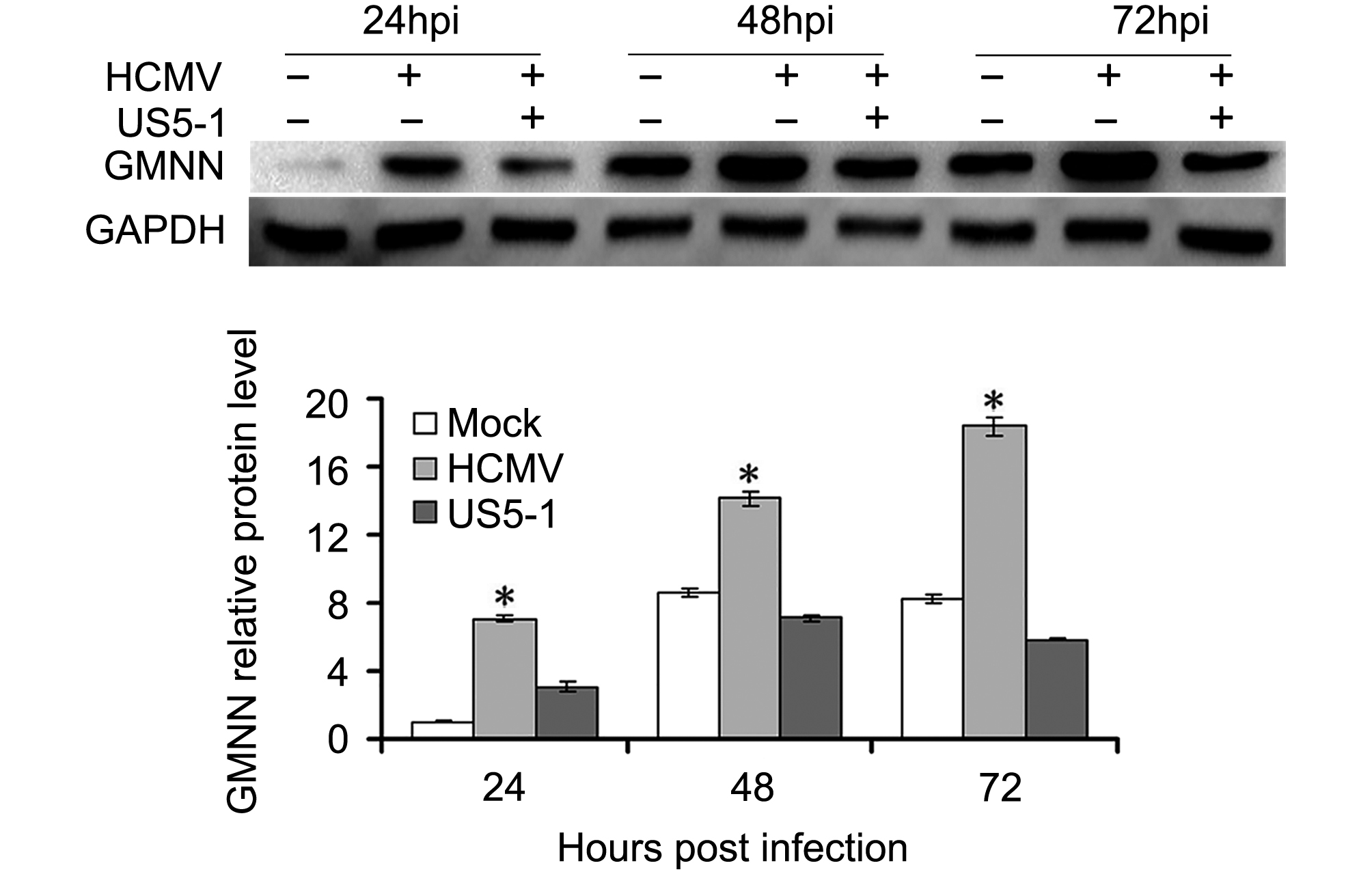
Figure 4. Reduction of GMNN by hcmv-miR-US5-1 during infection. (A) Expression levels of GMNN in U373 cells mock or infected by HCMV during natural infection or after transfection of hcmv-miR-US5-1 mimics were detected by western blot at 24, 48, 72 hpi. Data from three independent repetitions were used for statistical analysis. * indicates P < 0.05, by comparing to data from the mock infected and the hcmv-miR-US5-1 mimics transfected cells at the same time points.
To determine whether hcmv-miR-US5-1 could influence HCMV DNA replication by repressing GMNN, HCMV DNA copies as well as the expressions of hcmv-miR-US5-1 and GMNN were detected in U373 cells infected by HCMV after transfection of miRNA negative control, hcmv-miR-US5-1 mimics, hcmv-miR-US5-1 inhibitor or GMNN specific siRNA, respectively. As expected, transfection of hcmv-miR-US5-1 into U373 cells resulted in substantially increase of hcmv-miR-US5-1 expression compared to miRNA negative control-transfected cells, while, hcmv-miR-US5-1 expression was effectively blocked in hcmv-miR-US5-1 inhibitor-transfected cells (Figure 5A). Our data showed that GMNN was accumulated during HCMV infection in U373 cells (Figure 5B); meanwhile, hcmv-miR-US5-1 expression (Figure 5A) and HCMV copy number (Figure 5C) were also exhibited time-dependent increase. However, decrease of GMNN expression caused by hcmv-miR-US5-1 and GMNN siRNA (Figure 5B) markedly reduced HCMV DNA levels (Figure 5C). Moreover, hcmv-miR-US5-1 inhibitor mediated hcmv-miR-US5-1 silence in infected U373 cells increased GMNN expression at 24, 48, 72 hpi compared with miRNA negative control-transfected cells (Figure 5B), and the corresponding HCMV copy number was also increased (Figure 5C). These results indicated that hcmv-miR-US5-1 may reduce HCMV DNA replication by regulating expression kinetics of GMNN during HCMV infection.
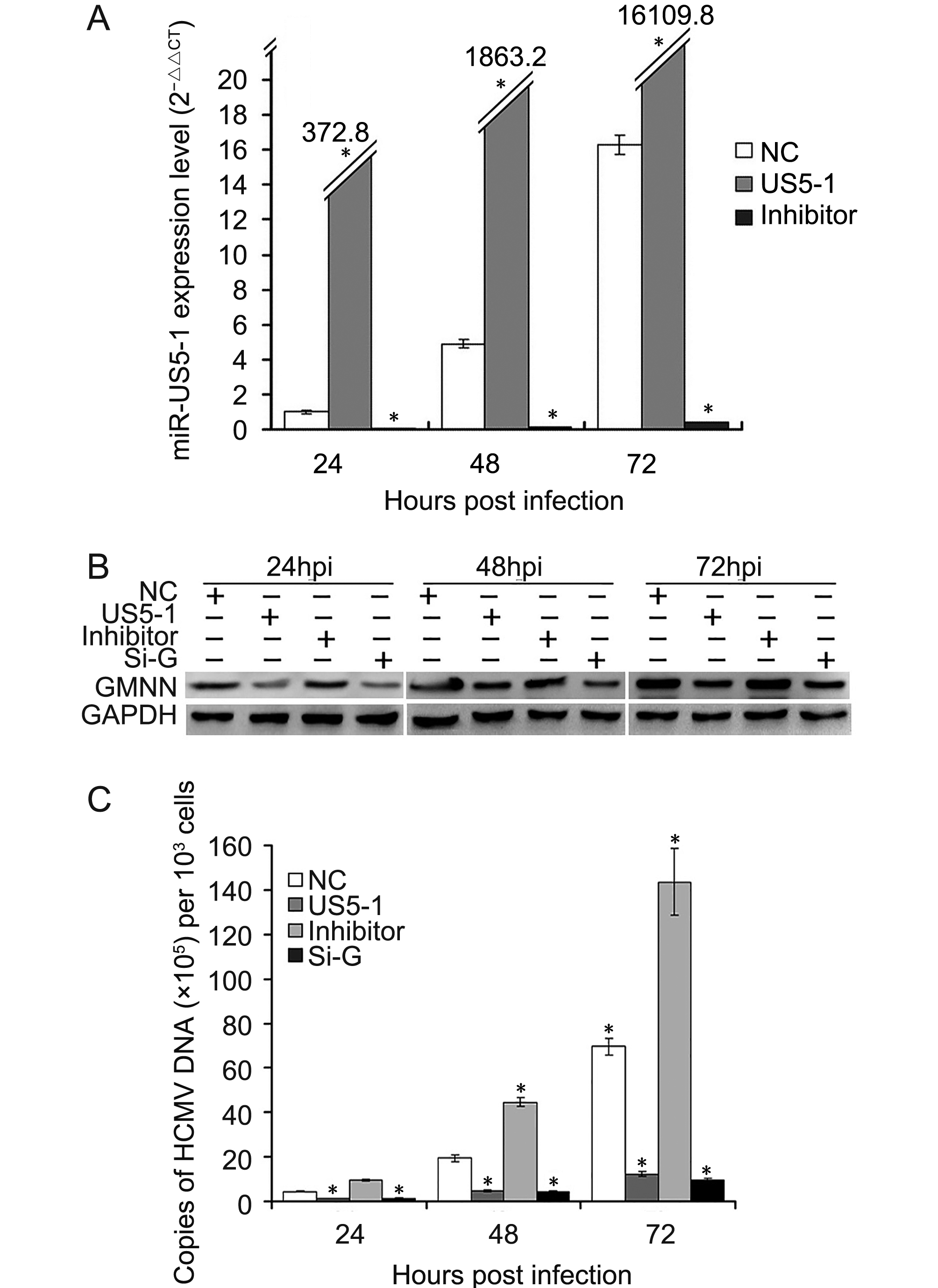
Figure 5. Reduction of HCMV DNA replication by hcmv-miR-US5-1 over-expression. U373 cells transfected with miRNA negative control (NC), hcmv-miR-US5-1 mimics (US5-1) or hcmv-miR-US5-1 inhibitor (Inhi) were infected with HCMV at 3 MOI 24 hours after transfection. At 24, 48, 72 hpi, expression levels of hcmv-miR-US5-1 were measured by RT-qPCR, and normalized to that of cells transfected with miRNA negative control at 24hpi. (B) U373 cells transfected with miRNA negative control, hcmv-miR-US5-1 mimics, hcmv-miR-US5-1 inhibitor or siRNA for GMNN (Si-G) were infected with HCMV at 0.5 MOI 24 hours after transfection. At 24, 48, 72 hpi, GMNN protein expressions were detected by western blot. (C) Viral DNA copies in cells treated as above were measured by real-time PCR at 24, 48, 72 hpi. The assays were performed in triplicate. Data from three independent repetitions were used for statistical analysis. * indicates P < 0.05 by comparing to data from miRNA negative control transfected cells at the same time points.
-
To determine whether decrease of endogenous GMNN protein level by hcmv-miR-US5-1 could influence the host cell cycle and proliferation, the cell cycle and cell number were measured as described above. As shown in Figure 6A, more cells progressed into the S and G2 cell cycle phases when transfected with hcmv-miR-US5-1 mimics or GMNN specific siRNA. Meanwhile, host cell proliferation also increased in these cells (Figure 6B). Silencing of hcmv-miR-US5-1 by its inhibitor counteracted these effects caused by the miRNA.
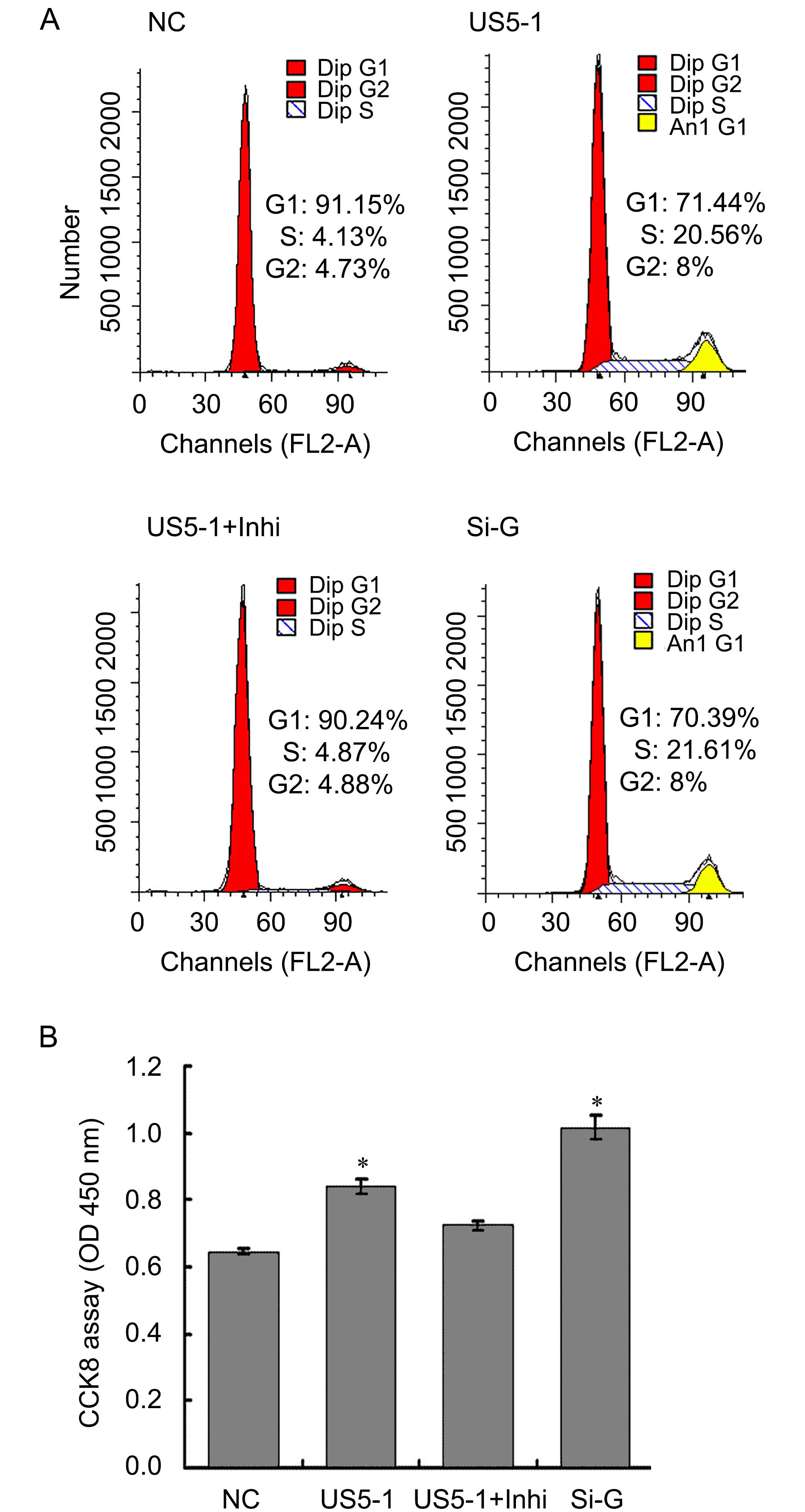
Figure 6. Modulation of host cell cycle and cell proliferation by hcmv-miR-US5-1 inhibiting GMNN expression. Cellular DNA contents of U373 cells transfected respectively with miRNA negative control (NC), hcmv-miR-US5-1 mimics (US5-1), hcmv-miR-US5-1 mimics and its inhibitor (US5-1+Inhi) or siRNA for GMNN (Si-G) were quantified by flow cytometry (FACS) analysis after stained with propidium iodide. (B) Cell proliferation was detected by CCK8 analysis. Data from three independent repetitions were used for statistical analysis. * indicates P < 0.05 by comparing to that of cells treated with miRNA negative control.
-
miRNAs are ubiquitously expressed among multicellular organisms. Several DNA viruses of the herpesvirus family including HCMV were also found to express miRNAs throughout their evolution (Gottwein and Cullen. 2008). The functions of HCMV encoded miRNAs may be more complicated than have been imaged, because they could potentially regulate both viral and host genes. It was reported that hcmv-miR-UL112-1 inhibited viral replication by targeting the major immediate early gene IE72, (UL123, IE1), UL112/113, UL120/121 and UL144 (Stern-Ginossar et al., 2009; Grey et al., 2007). Meanwhile, hcmv-miR-UL112-1 can also reduce the expression of major histocompatibility complex class I-related chain B (MICB) and interleukin-32 (IL-32), which may benefit for viral immune evasion (Huang et al, 2014; Stern-Ginossar et al., 2007). Moreover, hcmv-miR-US25-2 can reduce HCMV replication by targeting IE72, pp65 expression and eukaryotic translation initiation factor 4A1 (eIF4A1) (Qi et al., 2013), while hcmv-miR-US33 can inhibit viral DNA synthesis by down-regulating expression of the host Syntaxin3 (STX3) (Guo et al., 2015). In addition, hcmv-miR-US4-1 was reported to inhibit CD8+ T cell responses by targeting the immunoevasion protein aminopeptidase ERAP1 (Kim et al., 2011). Furthermore, hcmv-miR-US25-1 was demonstrated to inhibit HCMV DNA replication through the reduction of IE72 and pp65 expression (Khoshnevis and Tyring, 2002), it was also reported to downregulate multiple cellular genes associated with cell cycle control, such as cyclin E2, BRCC3, EID1, MAPRE2 and CD147, by interacting within the 5′UTR of the targets (Grey et al., 2010). Recently, hcmv-miR-UL70-3p and hcmv-miR-UL148D was demonstrated to play important roles in overcoming the cellular apoptosis (Babu et al., 2014). These collected results suggest that HCMV encoded miRNAs target both viral and cellular genes in order to achieve immune evasion and regulate viral replication as well as diverse cellular processes such as cell cycle and cellular apoptosis.
Mature hcmv-miR-US5-1 is derived from the 3′ arm of the pre-hcmv-miR-US5-1 stem-loop structure encoded by the intergenic region between HCMV US6 and US7 (Grey et al., 2007; Stark et al., 2012). However, knowledge about the biological function of hcmv-miR-US5-1 is very limited. In present study, 23 putative targets of hcmv-miR-US5-1 were successfully identified using hybrid-PCR method. Among them, the cellular DNA replication inhibitor GMNN, which has been demonstrated to play a critical role in cell cycle regulation and accumulate during HCMV infection (Biswas et al., 2003), was identified as a direct target of hcmv-miR-US5-1. The binding site of hcmv-miR-US5-1 was located in GMNN coding sequence and validated by luciferase assay and western blot. Compared to controls, cotransfection of pMIR-GMNN with hcmv-miR-US5-1 mimics showed a significant lower luciferase activity (~25%). However, mutation of the hcmv-miR-US5-1 binding site in the GMNN coding sequence abolished this effect. Moreover, ~56.5% decrease of endogenous GMNN protein expression was observed in the presence of hcmv-miR-US5-1. It could be confirmed that hcmv-miR-US5-1 has the ability to specifically repress GMNN expression through the identified binding site in the CDS of GMNN.
GMNN was reported to accumulate during HCMV infection and subsequently inhibit the licensing of cellular DNA replication. In this study, the expression of GMNN was indeed increased in HCMV infected cells, and its level accumulated as the infection progressed. However, over-expression of hcmv-miR-US5-1 could reduce the endogenous protein expression of GMNN, and block the accumulation of GMNN during HCMV infection. It worth noting that, hcmv-miR-US5-1 exhibited time-dependent increase but didn’t block the accumulation of GMNN during natural HCMV infection. It can be assumed that, except for hcmv-miR-US5-1 which downregulate the expression of GMNN, there must be other undefined factors can stimulate the expression of GMNN in the process of natural HCMV infection.
As the cellular DNA replication inhibitor, GMNN can inhibit DNA replication by binding to DNA replication factor Cdt1. Since hcmv-miR-US5-1 could reduce the accumulation of GMNN during HCMV infection, it could subsequently stimulate host cell DNA synthesis, that competitively inhibiting viral replication. In this study, the HCMV DNA replication was also detected in hcmv-miR-US5-1 over-expressed cells. At different hours post HCMV infection, viral DNA copies in cells transfected with hcmv-miR-US5-1 was reduced compared with that in cells transfected with miRNA control. hcmv-miR-US5-1 inhibitor destructed this reduction. Additional, utilizing of GMNN specific siRNA ruled out possible off-target effects of hcmv-miR-US5-1 on HCMV DNA replication. This result indicated that over-expression of hcmv-miR-US5-1 can inhibit HCMV replication by specifically targeting GMNN. In our further study, we would like to detect multiple viral key regulators, such as IE proteins, to explore more possible mechanism that hcmv-miR-US5-1 might regulate viral replication.
GMNN, which plays a critical role in cell cycle regulation, has been also reported to play an important role in cell proliferation (Yoshida et al., 2004; Barry et al., 2012; de Renty et al., 2014). Previously reports have demonstrated that depletion of GMNN could promote cell cycle progression and increase host cell proliferation (Wohlschlegel et al., 2002; Shreeram et al., 2002; Klotz-Noack et al, 2012). As GMNN protein expression was significantly decreased in cells transfected with hcmv-miR-US5-1, it is interesting to know whether over-expression of hcmv-miR-US5-1 could also affect the host cell cycle and proliferation or not. Our results revealed that more cells transfected with hcmv-miR-US5-1 mimic or GMNN specific siRNA progressed into the S and G2 cell cycle phases, and host cell proliferation was also increased in these cells. Cells progressed into the S and G2 cell cycles are conducive to host cellular DNA synthesis. This process may consume large amounts of essential nutrients and cellular enzymes needed for viral DNA synthesis, thus competitively inhibiting viral DNA replication (Caffarelli et al., 2013; Fehr and Yu, 2013). We used miR-US5-1 inhibitor in the recovery test, and confirmed the changes of cell replication was due to the increase of miR-US5-1. Meanwhile, we noticed that an inhibitor alone group might be added in the experiment to measure its direct effect.
Based on the functions of GMNN and our studies above, it can be assumed that HCMV probably activate GMNN to facilitate viral replication but simultaneously inhibits host cell DNA synthesis by an undefined mechanism. However, over expression of hcmv-miR-US5-1 could probably inhibit viral replication and influence host cellular environment via blocking the accumulation of GMNN during HCMV infection. In natural infection, HCMV may accurately control the viral replication at post transcription level by downregulation of GMNN via hcmv-miR-US5-1. This may be one possible strategy that HCMV adopt to facilitate establishment or maintenance of latency, further studies are required to understand the biological function of hcmv-miR-US5-1 during HCMV infection.
In conclusion, our study identified that hcmv-miR-US5-1 could directly target the DNA replication inhibitor Geminin (GMNN) and inhibit virus replication by blocking the accumulation of GMNN during HCMV infection. Meanwhile, inhibition of GMNN caused by hcmv-miR-US5-1 could influence host cell cycle and proliferation. These findings provide a new insight into hcmv-miR-US5-1 biological function during HCMV infection.
-
This work was supported by the National Natural Science Foundation of China (81371788 and 81171580) and the Specialized Research Fund for the Doctoral Program of Higher Education (20112104110012) and the Outstanding Scientific Fund of Shengjing Hospital.
-
The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
-
QR and RH conceived and designed the experiments; SJJ, XG and YZS performed the experiments; YQ and ZYL analyzed the data; ZRS and YPM contributed reagents/materials/analysis tools; SJJ, QR, RH and YJH wrote the paper.
Supplementary Table S1 is available on the websites of Virologica Sinica. www.virosin.org; link.springer.com/journal/12250.
-

Table S1. Primer sequences used in plasmid construction
Human cytomegalovirus miR-US5-1 inhibits viral replication by targeting Geminin mRNA
- Received Date: 14 August 2017
- Accepted Date: 26 October 2017
- Published Date: 31 October 2017
Abstract: Viruses commonly create favorable cellular conditions for their survival through multiple mechanisms. MicroRNAs (miRNAs), which function as post-transcriptional regulators, are utilized by human cytomegalovirus (HCMV) in its infection and pathogenesis. In the present study, the DNA replication inhibitor Geminin (GMNN) was identified to be a direct target of hcmv-miR-US5-1. Overexpression of hcmv-miR-US5-1 could block the accumulation of GMNN during HCMV infection, and the decrease of GMNN expression caused by hcmv-miR-US5-1 or GMNN specific siRNA reduced HCMV DNA copies in U373 cells. Meanwhile, ectopic expression of hcmv-miR-US5-1 and consequent lower expression of GMNN influenced host cell cycle and proliferation. These results imply that hcmv-miR-US5-1 may affect viral replication and host cellular environment by regulating expression kinetics of GMNN during HCMV infection.














 DownLoad:
DownLoad: