HTML
-
RNA modifications, which are abundant in cells, are regarded as tags for RNAs that are essential for gene expression (Engel and Chen 2018; Sanchez-Vasquez et al. 2018). N6-methyladenosine (m6A) is one of the most abundant modifications of RNAs, and especially messenger RNAs (mRNAs) (Gonzales-van Horn and Sarnow 2017; Ivanova et al. 2017; Shi et al. 2017). This modification expedites turnover of the transcript (Zhao et al. 2017). It can also regulate RNA fate including their transcription, translation (Wang et al. 2015), decay (Cao et al. 2016; Huang and Yin 2018), and involvement in biological processes (Shi et al. 2017) such as stem cell differentiation, embryonic development (Batista et al. 2014), and stress responses (Wu et al. 2017; Zhou et al. 2015).
In the cell, m6A methylation is affected by three proteins; it is installed by methyltransferases, which function as "writers", reversed by demethylases termed "erasers", and recognized by specific m6A-binding proteins that act as "readers" (Fig. 1) (Yue et al. 2015). The methyltransferase "writers" are methyltransferase-like protein 3 (METTL3), METTL14, the associated protein Wilms' tumor 1-associated protein (WTAP), and METTL16, recently identified by Warda et al. (Huang et al. 2018; Liu et al. 2014; Scholler et al. 2018; Warda et al. 2017). For many years, METTL3 was only regarded as an AdoMet-binding subunit that participates in mRNA modifications (Bokar et al. 1997), until METTL14 was identified. In mammalian cells, METTL3 and METTL14 form a heterodimer complex to regulate the m6A methylation of mRNA (Huang et al. 2018; Liu et al. 2014; Scholler et al. 2018); they add a methyl to the amidogen of adenosine, which occurs mainly in the 5' untranslated region (UTR) and 3' UTR near the stop codon, but can also be present in long exons (Gonzales-van Horn and Sarnow 2017). Interestingly, an mRNA with sites for m6A methylation in the 5'UTR can be translated in a cap-independent manner, directly binding eukaryotic initiation factor 3 (eIF3) without cap-binding factor eIF4E, a protein normally associated with translation initiation (Meyer et al. 2015). Further, the methylation of METTL3 and METTL14 can be reversed by demethylases such as fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5), which function as "erasers" and belong to the ALKB family (Aik et al. 2014; Zhu and Yi 2014). FTO prefers to bind intronic regions in the premRNA, and in particular sites near alternatively spliced exons. The m6A then forms two intermediates, namely N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (fm6A) (Fu et al. 2013), and the absence of FTO leads to substantial changes and exon skipping in the pre-mRNA (Bartosovic et al. 2017). ALKBH5 directly binds adenosines of m6A-modified mRNAs without any intermediate (Wu et al. 2017; Zheng et al. 2013). In addition, m6A modification is regulated by "readers" that contain the YTH domain, specifically the proteins YTHDF1-3, which all specifically bind m6A-modified mRNAs (Wu et al. 2017). YTHDF1 interacts with translation initiation factors to facilitate the formation of mRNA (Wu et al. 2017). YTHDF2 is also essential for the post-transcriptional regulation of mRNA (Ivanova et al. 2017) by directly recruiting the CCR4-NOT deadenylase complex to destabilize m6A-RNA, which leads to mRNA degradation (Du et al. 2016). YTHDF3 is also a "reader", but its function is not yet clear. Some studies indicate that YTHDF3 accelerates mRNA translation (Li et al. 2017a) together with YTHDF1 and facilitates the decay of m6A RNA (Shi et al. 2017). However, some studies show that YTHDF2 might play dual roles for RNAs. For example, YTHDF2 was found to enhance HIV-1 protein and RNA expression by promoting mRNA translation in HIV-producing-CD4+ cells (Kennedy et al. 2017; Lu et al. 2018), but it also suppresses HIV infection by degrading gRNA in HIV-1 target cells (Lu et al. 2018; Tirumuru et al. 2016). Furthermore, there are other "readers", termed YTHDC proteins, with a YTH domain, including YTHDC1-2, which can also recognize m6A sites through the preferential recognition of GG(m(6)A)C sequences in the nucleus (Xu et al. 2014).
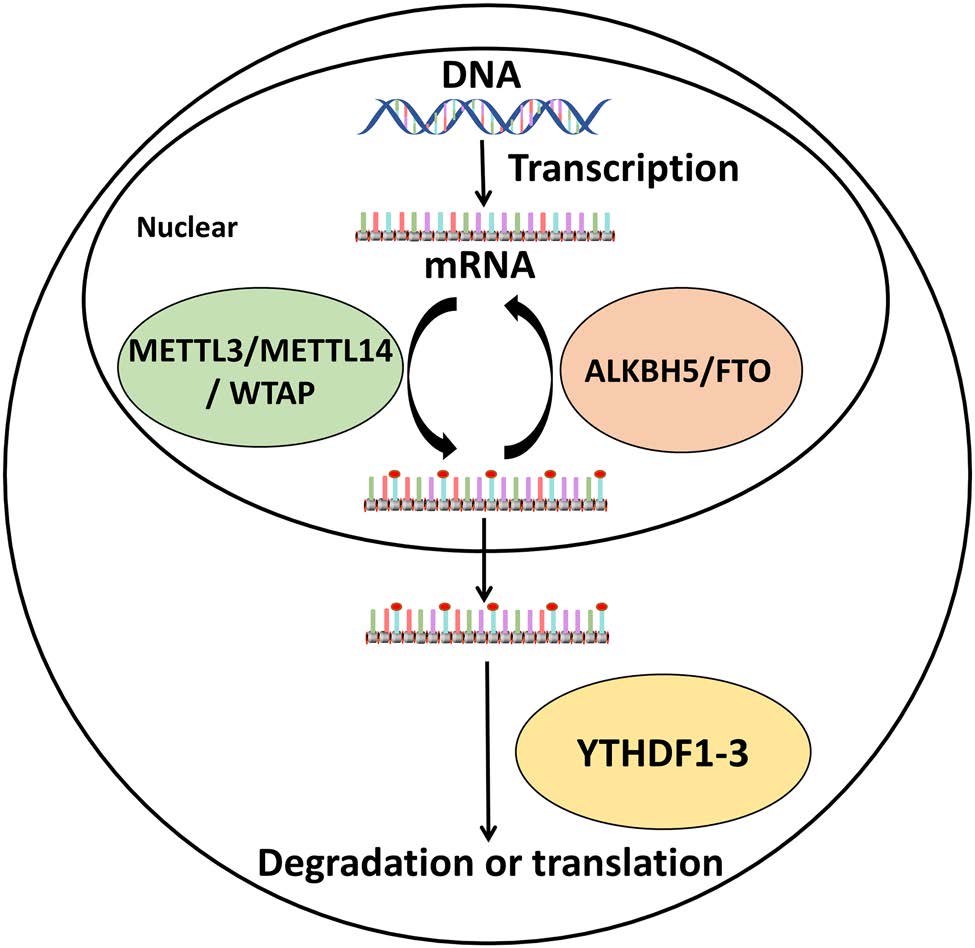
Figure 1. The machinery of m6A methylation. RNAs, especially mRNA, are methylated by the methyltransferases METTL3, METTL14 and WTAP, which is reversed by the demethylases FTO and ALKBH5. In addition, m6A-modified mRNAs can be recognized by YTHDF1-3, leading to degradation or translation.
Many scientists have focused their studies on the roles of N6-methyladenosine in vital processes including T cell functions, virus activities, and cancers, with the goal of devising new therapies. We summarize the recent achievements in this field that systematically elaborate upon the roles of N6-methyladenosine in such process.
-
In the 20th century, it was found that m6A modifications are widespread in viruses including influenza (Krug et al. 1976), vaccinia virus (Wei and Moss 1975), Rous sarcoma virus (Kane and Beemon 1985) and simian virus 40 (SV40) (Canaani et al. 1979). Recent studies indicate that m6A is also present in other viruses such as human immunodeficiency virus (HIV) (Kennedy et al. 2017; Lichinchi et al. 2016a; Lu et al. 2018; Tirumuru et al. 2016), herpesvirus (Hesser et al. 2018; Ye 2017; Ye et al. 2017), and Flaviviridae (Gokhale et al. 2016; Lichinchi et al. 2016b); there has also been much progress in studies on polyomavirus (Finkel and Groner 1983; Tsai et al. 2018) and influenza A virus (IAV) (Courtney et al. 2017).
-
Recently, three groups revealed the vital roles of m6A methylation in the HIV life cycle (Kennedy et al. 2017; Lichinchi et al. 2016a; Tirumuru et al. 2016). They confirmed that m6A methylation is beneficial for virus replication and viral gene expression; specifically, this was increased when they suppressed the demethylases FTO and ALKBH5 and was decreased when they inhibited the methyltransferases METTL3 and METTL14 (Kennedy et al. 2017; Lichinchi et al. 2016a; Tirumuru et al. 2016). Furthermore, the three teams identified the locations of m6A sites. Although they all mapped the sites to 5'UTRs, env/rev, and 3'UTRs, some differences were also noted among these publications. Kennedy et al. divided these sites into four m6A clusters containing two or three potential m6A methylation sites, including an env/rev cluster, a U3/NF-κB cluster, a transactivation response (TAR) cluster, and a Nef cluster. When using YTHDF proteins to bind m6A sites, there was another edited site in the Nef cluster in primary HIV-1 isolates BaL and JR-CSF (Kennedy et al. 2017). In contrast, Lichinichi et al. identified 14 distinct m6A methylation peaks located in splicing junctions, coding regions, and noncoding regions, including two in the Rev-responsive element (RRE) bound by the viral Rev protein in a structural region (Lichinchi et al. 2016a). Tirumuru et al. identified multiple CLIP peaks of m6A sites that bind YTHDF1-3, covering TAR in the 5'UTR leader sequence, env/rev, and the 3'UTR (Tirumuru et al. 2016). Moreover, two of these publications showed that the abundance of m6A motifs changed after HIV-1 infection. Lichinichi et al. demonstrated that the frequency of the MGACK (A/C-GAC-G/U) motif was increased in infected T cells, and that the UGAC motif and MGACK motif were then preferred in HIV-1 RNA (Lichinchi et al. 2016a). Tirumuru et al. reported a slight increase in RRACH motif enrichment and a decrease in the GGACU motif in infected primary CD4+ T cells. In two of these studies, the roles of YTHDF1-3 proteins were found to be very different. Kennedy et al. showed that overexpression of YTHDF proteins in CD4+ T cells increased virus replication and HIV-1 viral proteins and mRNA, and they concluded that post-transcriptional m6A editing and YTHDF1-3 proteins are positive regulators of HIV-1 infection (Kennedy et al. 2017). In contrast, Tirumuru et al. found that YTHDF1-3 were suppressors and decreased HIV-1 reverse transcription to inhibit HIV-1 infection in CD4+ T cells upon overexpression (Tirumuru et al. 2016). The results of Lichinichi et al. were similar to those of Kennedy et al., and these authors also discovered that m6A methylation modulates Rev protein interaction with RREs in RNA, and that the methylation of an A7883 site modified by m6A in the RRE bulge region perturbs HIV-1 replication and RNA nuclear export (Lichinchi et al. 2016a).
To explain the different roles of YTHDF1-3 based on the results of these different groups, Tirumuru et al. carried out further analysis using different cell lines, specifically HIV-1-target cell lines and viral processing cell lines. In the HIV-1-target cell lines such as the HeLa cells, YTHDF1-3 inhibited HIV-1 infection by decreasing levels of gRNA and preventing viral reverse transcription. In virus-processing cell lines such as HEK293T cells transfected with pNL4-3 or TZM-bl, YTHDF1-3 proteins promoted viral gag expression and virus production but suppressed HIV-1 infectivity (Fig. 2). Moreover, YTHDF1-3 proteins, which preferentially bind to m6A RNA, formed complexes with gag in the presence of HIV-1 m6A-RNA (Lu et al. 2018). Those results explain the different roles of YTHDF1-3 in different cell lines.
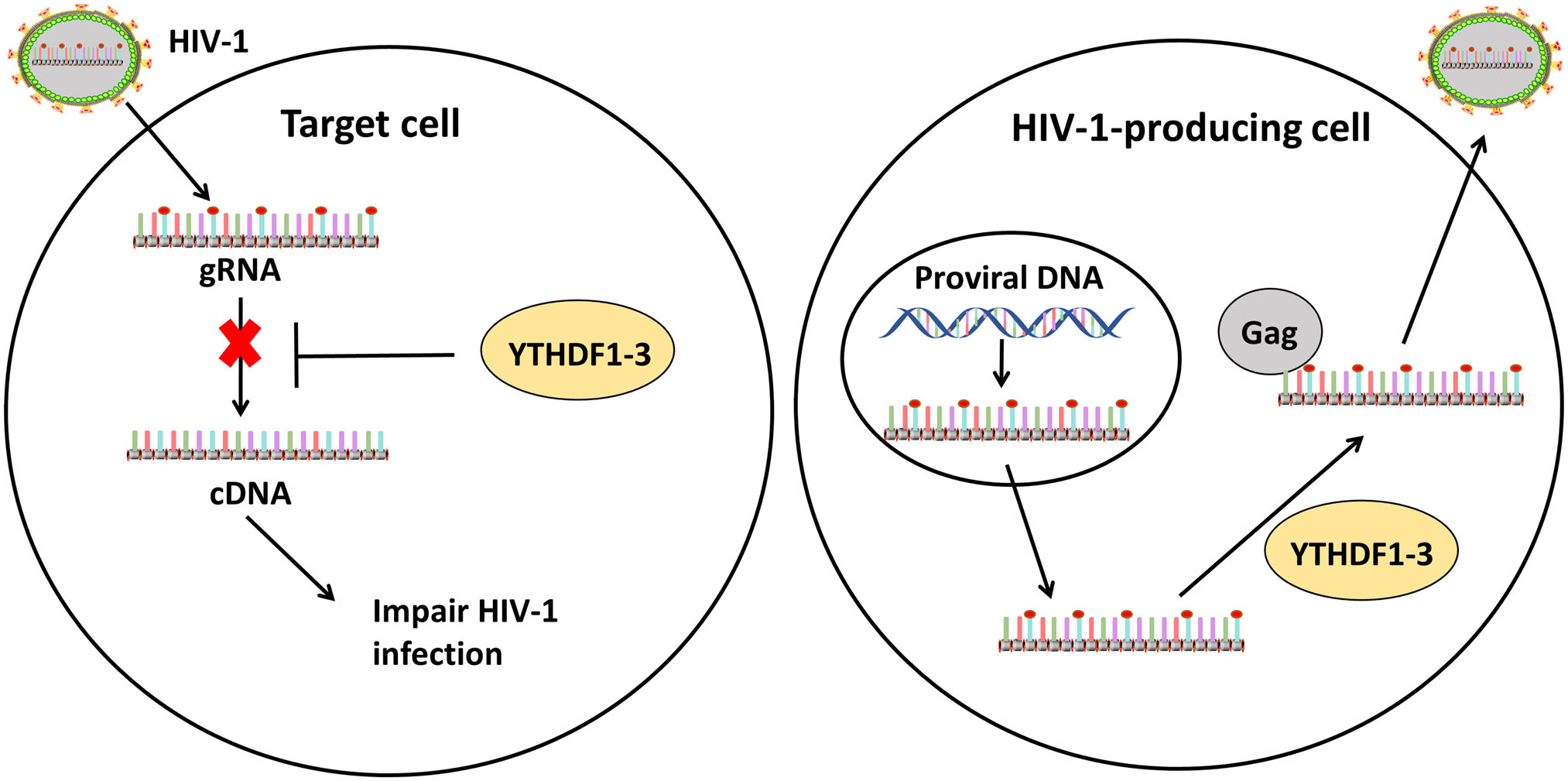
Figure 2. The roles of m6A methylation in HIV-1 life cycle. When HIV-1 viruses enter target cells, the exposed gRNAs, transcribed from the HIV-1 genome, are recognized by YTHDF1-3 proteins and degraded, which inhibits reverse transcription and virus infection (left). In HIV-1-processing cells, gRNAs are methylated and recognized by YTHDF1-3 proteins, which promotes HIV-1 Gag protein expression and virus production (right).
Moreover, in addition to different cells, the diverse reagents and experimental methods might explain the diametrically opposed roles of YTHDF proteins in virus replication. Kennedy et al. mainly focused on the transcription of viral RNA, whereas Tirumuru et al. paid more attention to the reverse transcription of viral RNA. The regulation of m6A on viral RNA in cells might thus be complicated. Despite the inconsistent results regarding the roles of m6A in regulating the replication of HIV virus and the expression of viral genes, all three publications indicated that m6A methylation plays an important role in HIV life cycle, suggesting that the methylation of viral genes might be a potential treatment target for AIDS patients, although the effectiveness of this approach needs to be verified.
-
Latency is a trait of all herpesviruses, and these viruses can be induced to enter productive lytic replication in the presence of some agents such as 12-O-tetradecanoylphorbol-13-acetate (TPA), tumor necrosis factor alpha (TNF-α), sodium butyrate, and hydrogen peroxide (H2O2) (Ye et al. 2017). Kaposi's sarcoma-associated herpesvirus (KSHV), a herpesvirus, is an oncogenic human deoxyribonucleic acid (DNA) virus. Three recent publications revealed that KSHV mRNA contains m6A modifications. In KSHV-infected cells, the levels of mRNAs modified by m6A methylation were increased when latent KSHV was stimulated to undergo lytic replication (Hesser et al. 2018; Ye 2017; Ye et al. 2017). Further, Ye et al. and Hesser et al. discovered that suppressing m6A by knocking down METTL3 inhibited splicing of the pre-mRNA encoding the replication transcription activator (RTA), which was important for the lytic replication of KSHV, with opposite results obtained after knocking down FTO (Hesser et al. 2018; Ye et al. 2017). Moreover, Ye et al. obtained a similar result by using the m6A catalytic reaction inhibitor 3-deazaadenosine or the FTO-selective inhibitor meclofenamic acid (Ye et al. 2017). Therefore, m6A methylation is favorable for lytic KSHV gene expression and replication. These two teams also revealed that the lytic switch protein RTA strongly induces m6A methylation and enhances its own pre-mRNA splicing (Hesser et al. 2018; Ye 2017; Ye et al. 2017). In different cells, they found that several m6A sites of open reading frame 50 (ORF50/RTA) are responsible for ORF50 (RTA) pre-mRNA splicing when KSHV was activated by TPA stimulation. They also found that the m6A sites of ORF50 (RTA) pre-mRNA are bound by YTHDC1 and splicing factors SRSF3 and SRSF10 in the nucleus (Ye 2017; Ye et al. 2017), whereas those sites are bound by YTHDF1-3 in the cytoplasm (Hesser et al. 2018). Ye et al. demonstrated that RTA can increase its own expression through transcriptional and post-transcriptional mechanisms and that KSHV has two opposite mechanisms to operate the host m6A machinery to regulate lytic replication and latency, respectively (Ye 2017; Ye et al. 2017). Hesser et al. showed that the m6A pathway controls ORF50 expression post-transcriptionally leading to a subsequent defect at the ORF50 promoter (Hesser et al. 2018).
The experiments of these two groups were performed using different cell lines, but they obtained similar results, specifically that m6A has a positive effect on the expression and replication of KSHV genes by regulating the splicing of RTA pre-mRNA. These results indicate that this epitranscriptomic modification might play a similar role in other herpesviruses. Therefore, understanding the role and regulatory mechanism of m6A modification in other herpesviruses could provide a new strategy to control herpesvirus infections.
-
Members of Flaviviridae, including Zika virus (ZIKV), dengue virus (DENV), West Nile virus, yellow fever virus (YFV), and hepatitis C virus (HCV), are positive singlestranded RNA viruses. Recent studies have shown that Flaviviridae viruses all contain m6A-modified sites, which regulate their gene expression and replication (Gokhale et al. 2016; Lichinchi et al. 2016b).
Gokhale et al. reported that m6A plays an important role in regulating the life cycle of HCV. Knocking out METTL3 and METTL14 increased the production of infectious HCV particles, and YTHDF1-3 proteins recognize and bind m6A sites of HCV RNA to suppress viral infection, indicating that m6A negatively regulates HCV. Then, this group mapped the m6A sites of the HCV RNA genome and found that m6A enhanced viral titer by increasing the interaction between the RNA and the core protein of HCV. Finally, they described several other Flaviviridae viral RNA m6A epitranscriptomic maps, including those of ZIKV, DENV, and YFV. Taken together, their findings revealed that m6A regulates viral infection and form the basis of future studies to explore the function of m6A within the broader Flaviviridae family of viruses (Gokhale et al. 2016).
ZIKV was discovered in 1947 (Driggers et al. 2016), and it can induce severe neurological defects. Lichinchi et al. reported that ZIKV infection alters the N6-methyladenosine topology and the function of viral and human RNAs. They identified 12 m6A peaks by performing methylated RNA immunoprecipitation-sequencing (MeRIP-seq) experiments, and these peaks were abundant in ZIKV RNA. Knockdown of METTL3/METTL14 or ALKBH5/FTO increased or decreased viral mRNA levels, respectively. Moreover, silencing YTHDF1-3 proteins, which were found to bind ZIKV viral RNA, inhibited replication. The authors also discovered that ZIKV infection influences RNA methylation sites of host cell transcripts, and that new m6A sites are preferentially deposited in the 5'UTR and coding sequence (Lichinchi et al. 2016b).
As described, these studies show that m6A methylation plays an important role in regulating Flaviviruses. The methyltransferases METTL3 and METTL14 can modify mRNA and inhibit viral replication in ZIKV and HCV. However, the enzymes FTO and ALKBH5 can suppress this effect and thus exert an adverse effect on the lifecycle of these viruses. In addition, YTHDF1-3 recognize m6A-modified sites to degrade viral mRNA. These studies showed that methylation-regulatory proteins in the host have inhibitory effects on Flaviviridae viruses, which provides a new direction for the clinical treatment of associated infections.
-
As early as the 1970s, m6A methylation was discovered in SV40, which belongs to polyomaviruses, a family of small DNA tumor viruses (Canaani et al. 1979; Finkel and Groner 1983; Tsai et al. 2018). The sites of m6A methylation in the mRNA of SV40 were found to occur in two sequences, specifically Gpm6ApC and (Ap)nm6ApC in late transcripts, which encode viral structural proteins (Canaani et al. 1979). In the 1980s, Finkel et al. confirmed that m6A methylation in pre-mRNAs is important for the formation of late SV40 mRNAs (Finkel and Groner 1983). Recently, Tsai et al. found that the addition of m6A to prototypic SV40 late mRNAs plays an important role in increasing viral gene expression and replication (Tsai et al. 2018). Tsai et al. mapped and identified 11 m6A sites in late transcripts and discovered that overexpression of the m6A-binding protein YTHDF2 led to more rapid viral replication and larger viral plaques, demonstrating that m6A plays a positive role in SV40 gene expression. Intriguingly, alternative splicing of SV40 was not found to be regulated by m6A. These results indicate that the addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication (Tsai et al. 2018).
-
IAV was first found to possess m6A modification of mRNAs in 1976, and each influenza viral mRNA species contains an average of three m6A residues (Krug et al. 1976). Narayan et al. determined that the distribution of m6A modifications in different influenza virus mRNAs is different (Narayan et al. 1987). Recently, Courtney et al. examined the regulatory mechanism of m6A modification with respect to IAV gene expression and replication. They demonstrated that IAV replication is inhibited by mutational inactivation of METTL3 in the human lung epithelial cell line A549, and that overexpression of YTHDF2 increases IAV replication and infectious particle production, which indicated that m6A methylation is advantageous for the IAV life cycle. However, not all RNAs including mRNA/cRNA (plus) strands and vRNA (minus) strands exhibited elevated m6A levels. Higher levels of m6A methylation addition to viral mRNAs encoding the structural proteins HA, NA, M1/M2, and NP and a lower level on the mRNAs encoding the viral polymerase proteins PB2, PB1, and PA were identified. Meanwhile, lower levels of HA mRNA and protein were observed in mutated IAV, whereas other IAV mRNAs and proteins were not changed (Courtney et al. 2017). These results show that m6A methylation regulates IAV gene replication and the expression of some proteins.
-
Recently, m6A modification to hepatitis B virus (HBV) RNA has been reported. Further, dual roles of m6A modification in the life cycle of HBV were demonstrated, as this was found to lead to decreased expression of HBV-related proteins, but enhanced the reverse transcription process of HBV pgRNA (Imam et al. 2018).
-
Meanwhile, some studies reported the function of m6A methylation in virus–host interactions. Kariko et al. found that unmodified RNA can stimulate the innate immune system through the activation of Toll-like receptors (TLRs), but that modified RNA including those subject to m6A methylation cannot active TLRs and trigger the immune defense system (Kariko et al. 2005). Similarly, retinoic acid-inducible gene Ⅰ (RIG-Ⅰ) can recognize RNA to activate innate immune signals, but it is not triggered by m6A-modified viral RNA, mediating viral evasion of the innate immune system (Durbin et al. 2016). In addition, DEAD-box (DDX) 46, a member of DDX family of helicases, binds RNA encoded by the antiviral genes Mavs, Traf3, and Traf6 in the nucleus and recruits the m6A demethylase ALKBH5 to demethylate these RNAs and retain them in the nucleus, thereby suppressing antiviral innate immunity in vivo during viral infection (Zheng et al. 2017). These results indicate that the m6A modification not only regulates the life cycle of the virus, but also exerts an effect on innate anti-viral immunity in the host. This provides a broader research direction for studying the effect of m6A modification on antiviral immunity.
In summary, m6A modification is important for the life cycle of the virus, and as m6A modification is more and more widely studied in different viruses, m6A modification could provide new therapeutic strategies to fight viral infection.
m6A Modification in Human Immunodeficiency Virus
m6A Modification in Herpesvirus
m6A Modification in Flaviviridae
m6A Modification in Polyomavirus
m6A Modification in Influenza A Virus
m6A Modification in Hepatitis B Virus
Host Factors Involved in m6A Modification
-
N6-methyladenosine on mRNA affects most post-transcriptional steps of gene expression and is associated with RNA metabolism including RNA stability, splicing, transport, translation, and localization (Brocard et al. 2017). m6A regulates alternative splicing by recruiting hnRNPC, a nuclear RNA-binding protein involved in premRNA processing (Liu et al. 2015). The "readers" can increase the stability of mRNA by reducing Hu antigen R binding (Brocard et al. 2017). Although m6A methylation has similar effects on mRNAs in different cell types, it still has different functions in diverse cells.
Li et al. demonstrated that T cell homeostasis is controlled by m6A mRNA methylation through the targeting of interleukin 7/signal transducer and activator of transcription 5/suppressor of cytokine signaling (IL-7/STAT5/SOCS) pathways, which regulate the homeostasis, differentiation, and proliferation of naïve T cells. STAT signaling can be inhibited by SOCS1, SOCS3, and CISH, members of the SOCS family, whose mRNAs are targeted by m6A. In wild-type naïve T cells, Socs1, Socs3, and Cish m6A mRNAs are recognized by YTHDF1-3 and degraded rapidly, and IL-7 signaling activated the Janus kinase (JAK)/STAT pathway to regulate the homeostasis, differentiation, and proliferation of naïve T cells (Fig. 3). In METTL3-deficient naïve T cells, Socs1, Socs3, and Cish mRNAs marked by m6A exhibited slower decay, leading to higher levels of these mRNAs and increased protein expression. Increasing activity of the SOCS family was found to suppress the IL-7/STAT signaling pathway and inhibit T cell homeostasis, differentiation, and proliferation. Additionally, the authors found that TH1 and TH17 cells were reduced and TH2 cells were increased in METTL3-deficient naïve T cell populations, but that the proportion of Treg cells was not changed compared to that in wild-type naïve T cells. However, m6A modification had no role in T cell apoptosis or TCR-mediated proliferation (Li et al. 2017b).
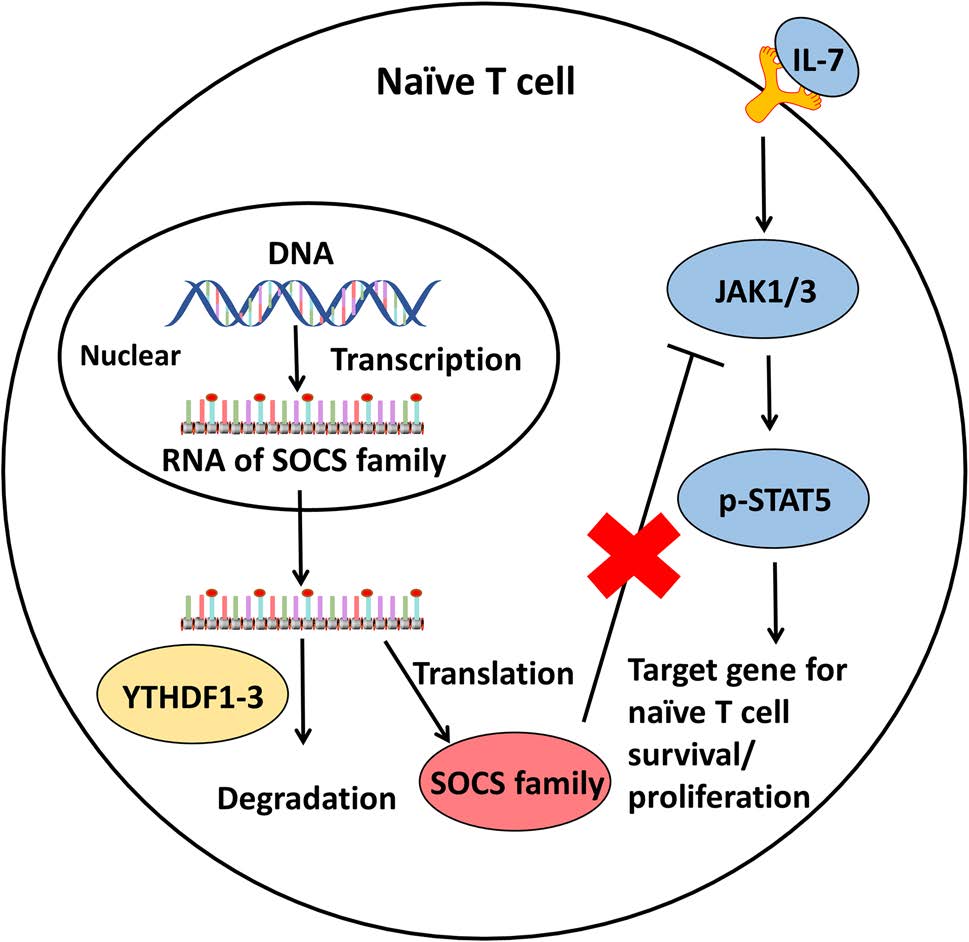
Figure 3. Regulation of m6A methylation in naïve T cells. The SOCS family inhibits the IL-7/STAT5/SOCS pathways to negatively regulate the homeostasis, differentiation, and proliferation of naïve T cells. However, RNAs of the SOCS family can be methylated in the nucleus, where they are recognized by YTHDF3 and degraded in the cytoplasm to sustain naïve T cell homeostasis, differentiation, and proliferation.
Meanwhile, Tong et al. revealed that m6A mRNA methylation has Treg-suppressive functions. By mapping m6A modifications, they found that these occur at a consensus sequence in the 3'UTR and 5'UTR in Treg cells. Depletion of METTL3 was found to increase mRNA levels of SOCS and suppress IL-2/STAT5 signaling, which is necessary for Treg functions and stability (Tong et al. 2018). These results showed that in different T cell subtypes, m6A RNA modification targets genes that encode essential components of signaling pathways that regulate the differentiation of naïve T cells, in addition to suppressing the functions of Tregs.
In addition, some studies have found that T cells are associated with the anti-viral mechanism of the host, and this is especially true for HIV (Che et al. 2010; Smith et al. 2016). The primary expansion of naïve T cells cultured with HIV-1-exposed dendritic cells was decreased and consequently immune dysfunction was shown to occur in vivo in HIV-1-infected individuals (Che et al. 2010); further, naïve CD8+ T cells were found to be required during persistent HIV-1 antiretroviral therapy (Smith et al. 2016). Moreover, TH1 cells can secret interleukin-21 and interferon-γ, which are necessary for maintaining the antiviral effect of IgG2 during IAV infection (Miyauchi et al. 2016). Thus, m6A methylation might indirectly regulate the host anti-viral effect by regulating naïve T cell homeostasis, differentiation, and proliferation, which might also provide new strategies for antiviral therapy, specifically through the regulation of naïve T cells.
-
N6-methyladenosine plays a vital role in virus replication and T cell homeostasis. During viral infection, m6A-modified mRNA levels are significantly elevated and therefore promote the replication and expression of viral genes. However, the function of YTHDF2 is different with different viruses, and some results obtained by different teams are conflicting due to the different cell lines and experimental methods used. Further, T cell homeostasis is controlled by m6A mRNA methylation, which targets the IL-7/STAT5/SOCS pathways.
This m6A methylation modification is present in viruses and regulates their life cycle. However, there are some questions that still need to be addressed. For viruses exhibiting latency, whether m6A methylation plays a role in latency or activation and how the virus hijacks the m6A machinery during infection are still unknown. Moreover, there are many viruses for which the roles of m6A methylation are still unknown. With advances in technology and the continuous improvement of experimental methods, these problems might be resolved in the future.
-
This study was supported by funding from the National Natural Science Foundation of China (Nos. 81672004, 31270202, 81801993, and 81801994), the Jilin University Science and Technology Innovative Research Team (JLUSTIRT, 2017TD-05), the Science and Technology Department of Jilin Province (20160101044JC), the Health and Family Planning Commission of Jilin Province (2013Z066), the Key Laboratory of Molecular Virology, Jilin Province (20102209), and China Postdocotoral Science Foundation (2018M63 1869).
-
The authors declare that they have no competing interests.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.














 DownLoad:
DownLoad: