HTML
-
Rotaviruses (RVs) are a major cause of acute gastroenteritis in infants and young children, claiming about 215, 000 lives worldwide annually (Tate et al. 2016). RVs are non-enveloped, double stranded RNA (dsRNA) viruses with 11 dsRNA segments encoding 12 proteins (Estes and Greenberg 2013). Each RV virion is composed of three concentric capsid layers. The outermost layer consists of the glycoprotein VP7 and the protease-sensitive spike protein VP4, which define the RV G and P genotypes, respectively (Matthijnssens et al. 2011). RVs are genetically diverse, consisting of at least 35 G and 50 P genotypes identified so far (https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg). VP4 can be cleaved by trypsin into two proteins, VP5* and VP8*, and VP8* interacts with cell surface carbohydrates for RV attachment (Fiore et al. 1991). Based on the VP8* sequences, the diverse P genotypes have been grouped into five genogroups (P[I]–P[V]) (Huang et al. 2012). P[4], P[6], P[8], and P[19] genotypes are grouped into P[II] genogroup, among which P[8] RVs are the most widely circulating human RVs (Yu et al. 2018). Several live-attenuated RV vaccines, including Rotateq® (Merck) and Rotarix® (GSK) that contain P[8] genotype, have been implemented in many countries, which has reduced the RV infection associated mortality and morbidity significantly (Rha et al. 2014; Burnett et al. 2018; Pindyck et al. 2018). Despite these advancements, the current vaccines do not show satisfactory efficacy in many developing countries and the reason for this low efficacy remain unknown (Desselberger 2017). Thus, further study for the better understanding of RV-host interaction, RV infection, and prevalence is necessary for improved control and prevention strategies to reduce mortality and morbidity caused by RVs.
Recognition of the cellular receptors is a crucial step in virus infection and is a major determinant of viral host range, tissue tropism, and epidemiology. VP8*, the RV carbohydrate-receptor binding protein, plays an important role in the initial cell attachment and recognizes glycan ligands in a genotype-dependent manner (Ruggeri and Greenberg 1991; Tan and Jiang 2014; Jiang et al. 2017). Some animal RVs were reported to be sialidase-sensitive previously, while human RVs were sialidase-insensitive (Ciarlet and Estes 1999; Kuhlenschmidt et al. 1999; Haselhorst et al. 2009). Histo-blood group antigens (HBGAs) have been demonstrated to be cell attachment factors for some human RVs based on structural and functional evidence (Hu et al. 2012; Huang et al. 2012; Liu et al. 2012), while mucin cores also interact with some human and animal RV VP8*s (Liu et al. 2016; Sun et al. 2016d; Pang et al. 2018). Both mucin cores and HBGAs are O-linked glycans that are synthesized by a group of glycosyltransferases. Four common core structures (core 1–4) constitute the majority O-glycans of mucins, such as human intestinal mucins that distribute abundantly on the epithelial cell surface, building up the intestinal mucus (Jensen et al. 2010). HBGAs are expressed on the red blood cells and epithelial cells at the mucosal surfaces and are also present in biological fluids, such as mucosal secretions, saliva, and milk. HBGAs are synthesized by sequential addition of monosaccharide to the disaccharide precursors with a β1, 3 and β1, 4 linkage and are highly polymorphic including the ABO, secretor (H), and Lewis families (Skovlund 1997; Tan and Jiang 2014), which may significantly affect RV epidemiology.
Several epidemiological studies have indicated an association between the polymorphic human HBGAs and susceptibility of RV infections. For example, human P[8] and P[4] RVs infected preferably Lewis positive secretors, while P[6] RVs could infect both Lewis positive and Lewis negative individuals (Nordgren et al. 2014; Van Trang et al. 2014). Further studies documented that non-secretors phenotype appear to be associated with resistance to severe gastroenteritis caused by P[8] RVs (Imbert-Marcille et al. 2014; Gunaydin et al. 2016; Zhang et al. 2016; Pollock et al. 2018). However, P[8] RV infection was also found in non-secretor individuals (Ayouni et al. 2015; Sun et al. 2016b). A recent study showed that the polymorphism of human HBGAs was associated with susceptibility to RVs. Nonetheless, in vitro infection of transformed cells appeared to be independent of HBGA expression (Barbe et al. 2018). These scenarios indicate the complexity in the interactions between RVs and various glycans, as well as the complex roles of HBGAs in human RVs infection, which needs to be further explored.
Human P[14], P[9], and P[25] RVs in the P[III] genogroup have been demonstrated to interact with A type HBGA (Hu et al. 2012; Liu et al. 2012), while human P[11] RVs in the P[IV] genogroup recognized type 1 and type 2 precursors (Liu et al. 2013; Ramani et al. 2013). Human P[4] and P[8] RVs were shown previously to interact with H1 and Lewis b antigens (Huang et al. 2012; Ma et al. 2015). Later, Liu et al. (2016) reported that VP8* of P[19] RV in the same P[II] genogroup bound to mucin core 2/4/6 and type 1 glycans. They also found that human P[4] and P[8] recognized mucin core 2, while human P[6], another P[II] genotype, did not bind to mucin core 2 but interacted with H type 1 precursor glycan lactose-Ntetrarose (LNT) (Liu et al. 2016). In addition, a study using nuclear magnetic resonance (NMR) spectroscopy indicated interaction between human P[8] RV (Wa strain) VP8* and GM1/GD1a (Haselhorst et al. 2009) and Wa RV was reported to recognize GM1 ganglioside during cellular infection (Fleming et al. 2014). Another study using saturation transfer difference (STD) NMR spectroscopy showed that type A HBGAs may be receptors for human P[4] RV DS-1 strain and P[6] RV-3 strain, but not P[8] Wa strain (Bohm et al. 2015). Thus, the RV-glycan interactions are complicated, particularly how the most prevalent P[8] RVs interact with the glycan ligands remains elusive.
Crystallography is a powerful tool to illustrate the detailed interactions and structural basis between RV VP8* and its glycan receptors. The crystal structure of P[14] HAL1166 VP8* complexed with type A-HBGA showed that the A-antigen binding site shares the same location as the sialic acid binding site in a Rhesus P[3] RV VP8* at one corner of the cleft region between the two b-sheets (bH and bK) (Dormitzer et al. 2002; Hu et al. 2012). The crystal structure of human P[11] N155 VP8* in complex with lactose-N-tetrarose (LNT) or lactose-N-neotetrarose (LNnT) revealed a larger glycan binding pocket beside the one of P[14] HAL1166, spanning almost the entire length of the cleft (Hu et al. 2015). The glycan binding pocket in P[19] appears to be unique compared with the other known ones, being located adjacent to the cleft and formed mainly by a 169–172 loop and a 209–212 loop (Liu et al. 2017; Sun et al. 2018). The crystal structures of human P[4] Indian and P[6] RV3 VP8*s complexed with type 1 glycan Lacto-N-fucopentaose 1 (LNFP1) have been solved recently, revealing the same glycan binding sites as that of P[19] VP8* (Hu et al. 2018). The crystal structures of P[8] Wa and Rotateq VP8* alone have been determined previously (Blanchard et al. 2007; Sun et al. 2016a). In this study, we further characterized the glycan binding specificity of P[8] RV VP8*s and determined the crystal structures of Rotateq P[8] VP8* complexed with mucin core glycan and H type 1 antigen, providing a structural basis for the P[8] RV-glycan ligand interactions.
-
VP8* genes (residues 1–231) of Rotateq P[8] (GenBank: GU565044), P[8]BEL(GenBank: JN849151) were synthesized in Genewiz (Suzhou, China) and cloned into the pGEX4T-1 vector. Human P[6] (5311142) (GenBank: KT162986), human P[4] (11151099) (GenBank: KT162987), human P[8]lab (11221075) (GenBank: KT162984) were described previously (Ma et al. 2015). Recombinant VP8* proteins with N-terminal GST-tag were expressed in E. coli BL21 (DE3) cells (Tiangen) as previously described (Sun et al. 2016a). GST-fusion VP8*s were purified with Glutathione beads (GE healthcare) using established procedures (Sun et al. 2016a). VP8* core (residues 64-223) fragment of Rotateq P[8] was cloned into the pET30a vector and expressed in E.coli with C-terminal his-tag. VP8* core protein was purified with HiTrap Fast Flow (GE healthcare). Further purification was performed with gel filtration using superdex 200 column with 20 mmol/L Tris–HCl (pH 8.0), 50 mmol/L NaCl. The concentration of the purified protein was determined by the BCA Kit (BD Biosciences) with 2 mg/mL BSA as a standard.
-
Oligosaccharide binding assay was carried out as previously reported (Sun et al. 2016a). GST fusion VP8* proteins were coated on the 96 well plate (Costar) at 100 μg/well at 4 ℃ overnight. After blocking with 5% non-fat milk, PAA-biotin labeled oligosaccharides (including mucin core 2, mucin core 4, mucin core 6, LNT, H type 1, H type 2, A, B, Lewis a, Lewis b) were added at two concentrations 0.2 and 1 μg/well respectively. The plate was left at 4 ℃ overnight. Horseradish peroxidase-conjugated streptavidin (Abcam) was then added at 0.1 μg/well after washing the plates 5 times with phosphate-buffered saline (PBS)-0.05% Tween 20. The reactions were conducted using a 3, 30, 5, 50- tetramethylbenzidine kit (BD Biosciences) and the absorbance of 450 nm was measured. The experiment was repeated twice and samples were performed with duplicates each time.
-
The Rotateq P[8] VP8* core protein in buffer (20 mmol/L Tris-HCl, 50 mmol/L NaCl, pH 8.0) was concentrated to approximately 20 mg/mL and crystallized using the sittingdrop vapor diffusion method at 18 ℃ with 1 μL of protein mixed with 1 μL of reservoir solution. Crystals were grown under the condition of 0.1 mol/L sodium acetate trihydrate pH 4.5, 2.0 mol/L ammonium sulfate. In order to obtain the protein-glycan complex, the glycans core 2 or LNFP1 were co-crystallized with the VP8* protein. The threoninelinked core 2 trisaccharide was prepared from Fmoc-protected peracetylated core 2-Thr (Sussex Research Laboratories, Ottawa, Canada) after deprotection (Johannes et al. 2015). Core 2-Thr was purified by high-performance liquid chromatography (HPLC) and analyzed by electrospray ionization mass spectrometry (ESI-MS) before use. The pentasaccharide Lacto-N-fucopentaose 1 (LNFP1) was purchased from Dextra Laboratories (UK). VP8* and core 2/LNFP1 were co-crystallized using a protein/ligand ratio of 1:50/1:200, respectively, under the same conditions as for the native crystal described above.
-
Crystals were dipped briefly in cryoprotectant solution containing 20% (v/v) glycerol and then flash-frozen in liquid nitrogen. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility BL17U. Data were processed using HKL2000 software (Otwinowski and Minor 1997). The structures were solved by molecular replacement using Phaser software (Read 2001) with the structure of Rotateq P[8] VP8* (Protein Data Bank [PDB] ID: 5JDB) as the search model (Sun et al. 2016a). The initial model was refined using REFMAC5, COOT, and PHENIX software (Emsley and Cowtan 2004; Adams et al. 2010). The structural analysis was carried out with the PyMOL software package (https://pymol.org/2/). The statistical data for P[8] VP8*-core 2 and VP8*-LNFP1 are presented in Table 1. Protein structure accession numbers: the structures of P[8] VP8*-core 2 and VP8*-LNFP1 have been deposited in PDB with the access codes of 6K2N and 6K2O, respectively.
Parametera RotaTeq P[8] VP8*-Core 2 RotaTeq P[8] VP8*-LNFP1 Data collection Space group P21 P6322 Cell dimensions a, b, c (Å) 66.289 80.454 114.55 80.454 73.789 116.593 α, β, γ (°) 90, 95.959, 90 90, 90, 120 Wavelength (Å) 0.97774 0.97852 Resolution (Å) 50.00-1.80 50.00-2.30 (1.86-1.8) (2.38-2.30) Rmerge (%)b 8.4 (80.4) 15.9 (86.0) I/σI 21.597 (1.500) 22.524 (4.889) Completeness (%) 98.4 (90.3) 99.57 (96.58) Redundancy 5.4 (5.4) 25.8 (27.0) Refinement Resolution (Å) 43.24-1.80 44.71-2.30 No. reflections 86599 10481 Rwork/Rfree 0.2024/0.2288 0.2035/0.2331 No. atoms Protein 7860 1336 Ligand/ion 80 52 Water 781 80 B-factors Protein 23.5 31.1 Ligand 59.8 74.7 Water 34.4 39.6 R.m.s. deviations Bond lengths (Å) 0.005 0.003 Bond angles (°) 0.69 0.54 Ramachandran plot Favored (%) 97.47 98.14 Allowed (%) 2.53 1.86 Disallowed (%) 0.00 0.00 aValues in parentheses are given for the highest resolution shell.
b Rmerge =$\Sigma \mathrm{hkl}|\mathrm{I}- <\mathrm{I}>| / \Sigma \mathrm{hklI} $ $ <\mathrm{I}>$ Table 1. Crystallographic X-ray diffraction and refinement statistics.
-
Human red blood cells (RBCs) of A, B, and O types and animal blood cells (Gelaidisi, Beijing) were washed twice with PBS before use and diluted to 1% RBCs. The GSTVP8* fusion proteins were twofold-serially diluted with 50 μL per well on 96-well V-bottom plates (Costar, Corning) and the original protein concentration was at 2 mg/mL. 50 μL of RBCs was added each well subsequently. The plate was incubated at 25 ℃ and agglutination was determined 1 h later. Wells without aggregates were considered as agglutination.
-
We performed fluorescent focus assays on African green monkey kidney epithelial cells (MA104 cells) in the presence and absence of glycans, with infections in the absence of glycans considered to be 100% infectivity. The MA104 cells were seeded on the 96-well plate (Costar). Laboratory-adapted Wa (G1P[8]) and SA11 (G3P[2]) rotavirus strains were investigated. Dilutions of virus yielding ~200 focus forming units per well were preincubated for 30 min with media containing human milk, LNFP1, mucin core 2, lactose, or media without glycan. Human milk was used at 1:10. The glycans were tested at a final concentration of 1 mg/mL and 5 mg/mL (for LNFP1). The MA104 cells on the 96-well plate were washed twice with ice-cold serum-free 1640 medium. After cooling the cells and virus to 4 ℃, the virus-milk or virus-glycan media were added and incubated on ice for 1 h. The inoculum was removed and cells were washed twice with ice-cold serum-free 1640 medium. The plates with serumfree 1640 medium were then put back in the incubator at 37 ℃ for 20 h. The cells were then fixed with ice cold methanol. Infected cells were detected with an anti-P[8] VP8* polyclonal rabbit serum (raised in-house, 1:400 dilution), followed by a fluorescein isothiocyanate (FITC)- conjugated goat anti-rabbit secondary antibody (Abcam, 1:1000 dilution).
-
Data analysis was performed using SPSS software. Analysis of variance (ANOVA) with Dunnett's correction for multiple comparisons was used. Values of P < 0.05 were considered to indicate statistical significance.
VP8* Proteins Expression and Purification
ELISA-Based Oligosaccharide Binding Assay
Crystallization
Data Collection, Processing, and Structure Determination
Hemagglutination Assay
Virus Infection Inhibition Assay
Statistical Analysis
-
In order to define the interactions between P[8] RV VP8*s and various glycans (Fig. 1A), VP8* proteins of three different P[8] RV strains, including the vaccine strain of Rotateq, the predominant strain in China in 2014, referred as P[8] 11221075, and another P[8] strain in 2009 BEL were produced for glycan binding assays (Fig. 1B). All these P[8] VP8* proteins bound conclusively to mucin core 2 glycans (Fig. 1C, 1D). P[8] 11221075 VP8* showed weak binding signals to mucin core 4 and 6 oligosaccharides (Fig. 1C). P[8] 11221075 VP8* displayed obvious binding to H type 1 glycans. P[8] VP8*s bound to mucin core 2 and H type 1 glycans in a dose-dependent manner (Fig. 1D). In addition, human P[4] 11151099 and P[6] 5311142 VP8*s also bound mucin core 2 glycans. Human P[8] and P[4] VP8*s appeared not to bind LNT, while human P[6] VP8* did (Fig. 1C). No interaction was seen between P[8]/P[4] VP8*s and type A, B, Lea, Leb, and H type 2 antigens (Fig. 1C). As a positive control, P[14] VP8* bound A antigen.
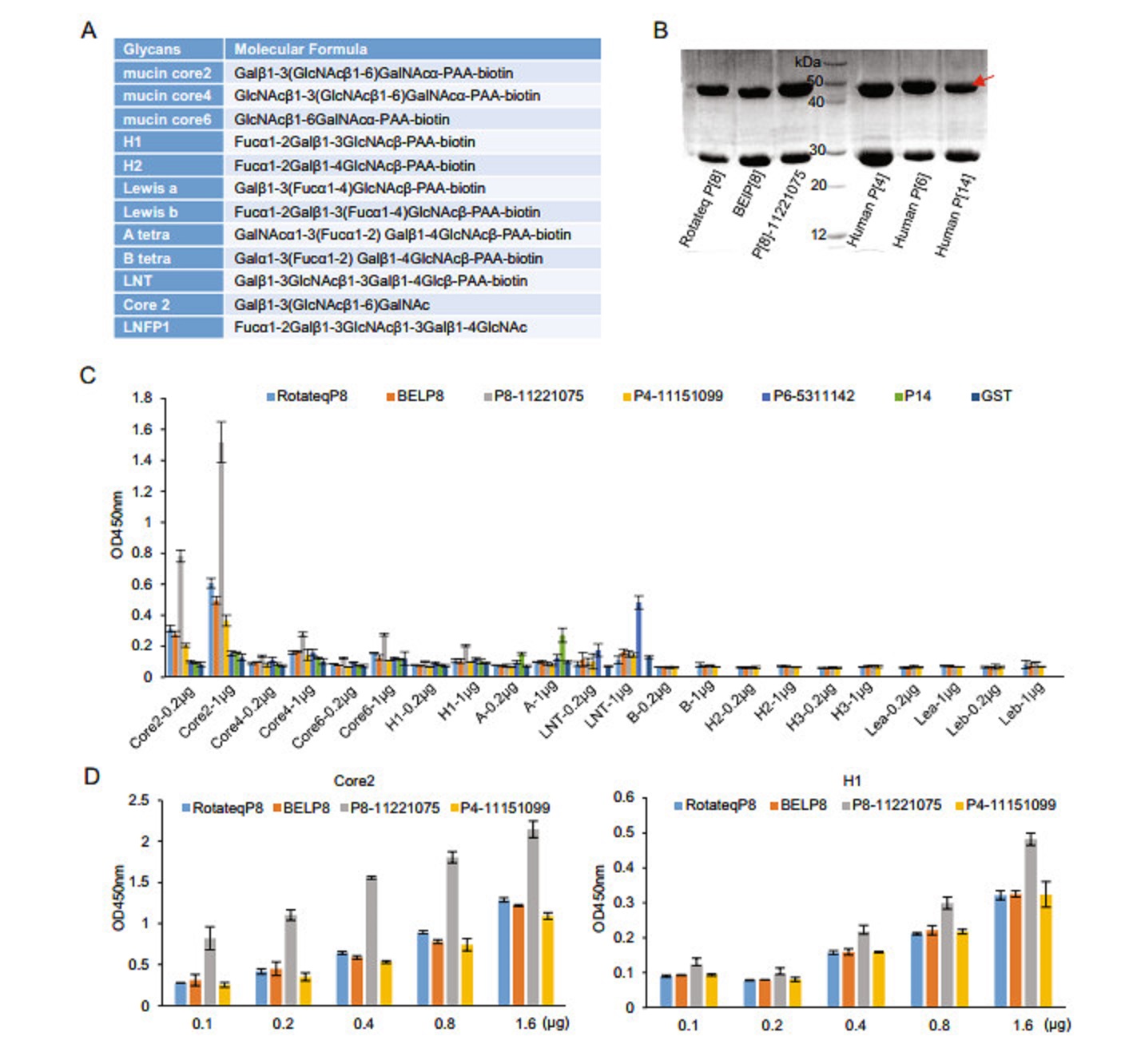
Figure 1. Glycan binding assay of P[8] VP8*s. A The glycans involved in this study are listed. B The SDS-PAGE of the GST-VP8* fusion proteins (~52 kDa). Red arrow indicates the proteins of interest. The 26 kDa protein was free GST. C GST-VP8* proteins of Rotateq P[8], BElP[8], labP[8]-11221075, human P[4]-11151099, human P[6]- 5311142, human P[14] were tested for the binding to oligosaccharides including core 2, core 4, core 6, H1, A, LNT (lactose-N-tetrarose, Galβ1-3GlcNAcb1-3Galβ1-4Glc) at two concentrations (0.2 lg and 1 μg). Interactions with B, H1, H2, lewis a, lewis b were also performed for Rotateq P[8], BElP[8], labP[8]-11221075, and human P[4]-11151099. Human P[14] bound to A-HBGA was a positive control and GST protein was used as the negative control. D Rotateq P[8], BElP[8], P[8]-11221075, and human P[4]-11151099 bound to core 2 and H1 with a series of glycan concentrations (0.1, 0.2, 0.4, 0.8, 1.6 μg).
-
The crystal structure of Rotateq P[8] VP8* in complex with a core 2 trisaccharide was determined at the resolution of 1.8 Å ngström (Å) (Fig. 2A). There were six protein molecules in the asymmetric unit. The electron density of core 2 were observed clearly in chain B and chain D. The core 2 trisaccharide of chain D was presented at 1 sigma level in the simulated annealing omit map (2mFo-DFc) (Fig. 2B). The residues W81, L167, Y169, G170, G171, R172, W174, T185, R209, and E212 of the VP8* protein participated in the interactions with the core 2 trisaccharide and thus build the cavity shaped glycan binding site (Fig. 2C). The edge of the glycan binding cavity is composed of the β-strand K, the I-J loop, and the 210 loop (residues 209–212), which links bstrand M and a-helix A; while the base of the binding cavity was formed by residues W81, L167, and W174. The core 2 trisaccharide was stabilized by a network of hydrogen bonds and hydrophobic interactions (Fig. 2C). The GlcNAc is the major binding saccharide forming four hydrogen bonds with T185, R209, E212, and four hydrophobic interactions with W81, L167, W174, T185, E212. In addition, the GalNAc formed two hydrogen bonds with R172and two hydrophobic interactions with G170 and R172, while the Gal only formed hydrophobic interactions with Y169, G170, and G171.
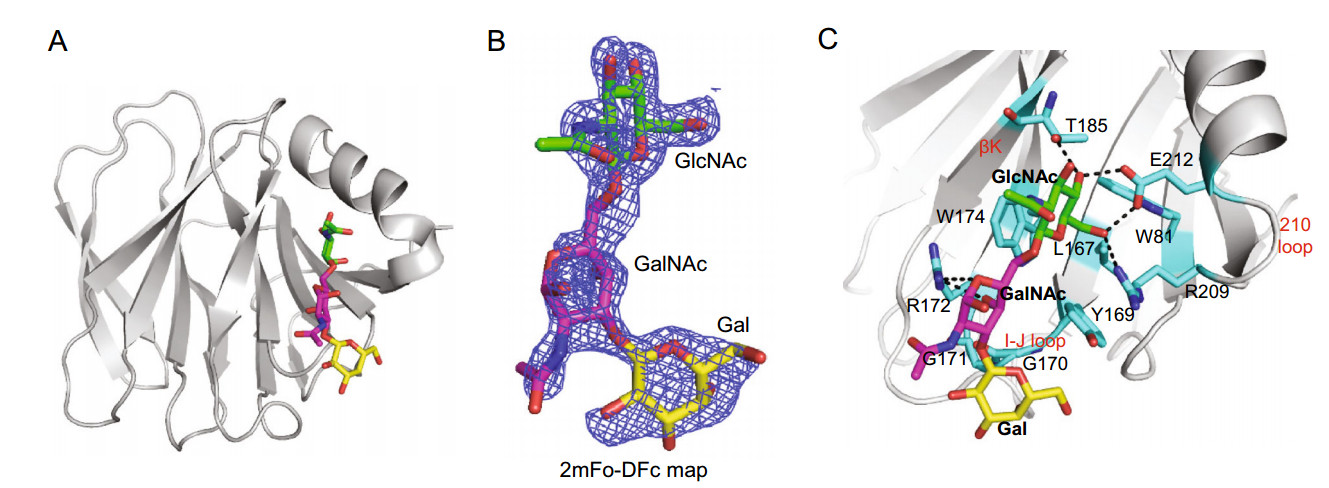
Figure 2. Crystal structures of Rotateq P[8] VP8*-core 2 complex. A The cartoon representation of P[8] VP8*-core 2 complex structures. The monosaccharide residues of core 2 are shown in stick representation. B The 2mFo-DFc omit difference electron density map of core 2 in P[8] VP8* structure, contoured at 1σ level is shown. GlcNAc, green; GalNAc, magenta; Gal, yellow. C The detailed interactions between core 2 and VP8*. The monosaccharide residues of core 2 and amino acids involved in the interaction are shown in stick representation. The hydrogen bonds are presented with dashed lines. The interactions were searched for by CCP4 (http://ccp4.ac.uk/) with the distance of less than 5 Å and then calculated by PyMOL (https://pymol.org/2/).
-
We also co-crystallized Rotateq VP8* in complex with an H type Ⅰ pentasaccharide, Lacto-N-fucopentaose 1 (LNFP1, Fuca1-2Galβ1-3GlcNAcb1-3Galβ1-4Glc) (Fig. 3A). The complex structure was determined at 2.3 Å resolution. There was one protein molecule in the asymmetric unit. The non-reducing end of LNFP1 was clearly seen at 1 sigma level of the electron density 2mFo-Fc map whereas the Gal moiety at the reducing end was not well-defined. Meantime, the Glc moiety at the reducing end were not shown because of the low map density (Fig. 3B). LNFP1 bound the VP8* protein at the same site as core 2 glycan. The residues W81, L167, Y169, G170, R172, W174, T184, T185, R209, E212, and N216 were involved in the interactions (Fig. 3C). The GlcNAcIII contributed mostly to the interactions by forming four hydrogen bonds with T185, R209, E212 and four hydrophobic interactions with W81, L167, W174, E212. The GalII at the non-reducing end interacted with T185 via a hydrogen bond and T184, T185, N216 through hydrophobic interactions, while Fuc-I was not involved in direct interactions. The GalIV at the reducing end further stabilized the glycan by forming three hydrophobic interactions with Y169, G170, and R172.
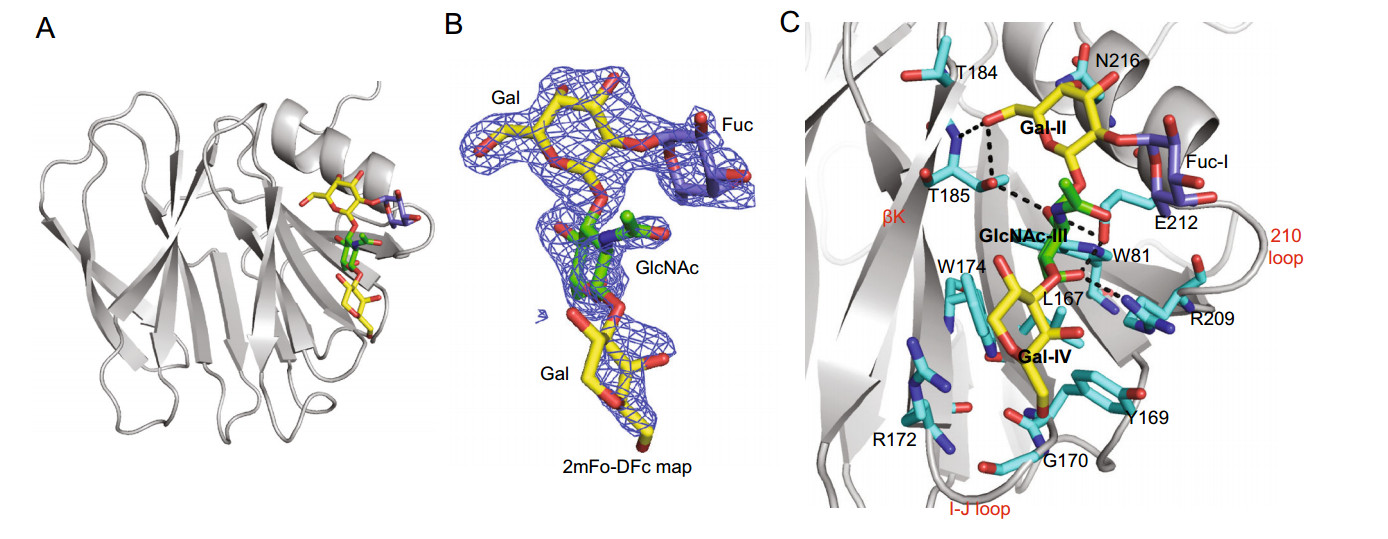
Figure 3. Crystal structures of Rotateq P[8] VP8*-LNFP1 complex. A The cartoon representation of P[8] VP8*-LNFP1 complex structures. LNFP1 is shown in stick representation. B The 2mFo-DFc omit difference electron density map of LNFP1 in P[8] VP8* structure, contoured at 1σ level is shown. GlcNAc, green; Gal, yellow; Fuc, slate. C The detailed interactions between LNFP1 and VP8*. The hydrogen bonds are presented with dashed lines. The interactions were analyzed in the same way as described in the legend of Fig. 2C.
-
Compared with the Rotateq VP8* structure (Sun et al. 2016a), glycan binding did not cause apparent conformational change, with the root mean squared deviation (RMSD) for alpha carbons of the backbone atoms between the bound VP8* and the free VP8* being 0.15 Å (mucin core 2) and 0.20 Å (LNFP I), respectively. Structural superimposition indicated that the core 2 trisaccharide and the LNFP1 pentasaccharide bound the same site of P[8] VP8*. There is a slight shift in the orientation of the GlcNAc moiety (pointed by a white arrow in Fig. 4A) between the bound mucin core 2 and the LNFP1 oligosaccharides, resulting in an orientation shift of the two glycan backbones within the binding cleft (Fig. 4A). In both cases, VP8* interacted with the glycans in a similar mechanism with the GlcNAc as the major interacting saccharide (Fig. 4B). Compared with the mucin core 2 trisaccharide, the LNFP1 pentasaccharide interacts with two additional residues, T184 and N216 of P[8] VP8* (Fig. 4B, 4C).
Structural analysis of the Rotateq P[8] VP8*-core 2 complex showed similar interaction mechanism as that seen in P[19] VP8*. Core 2 glycans lay at the same position of P[8] and P[19] VP8*s and interacted with the same set of amino acids except the residue 169 (Fig. 5A). Y169 in RotateqP[8] VP8* contributed a single hydrophobic interaction with Gal of the core 2 trisaccharide, whereas H169 in P[19] VP8* form two hydrophobic interactions with both GlcNAc and Gal (Fig. 5A).

Figure 4. Structural comparison of Rotateq P[8] VP8*-glycan structures. A Structure superimposition of Rotateq P[8] VP8*-core 2 and P[8] VP8*-LNFP1. Core 2/LNFP1 are shown in stick representation and colored green/yellow, respectively. The binding cavity is colored blue in the surface representation. The orientation of GlcNAc is highlighted by a white arrow. B Detailed presentation of the P[8] VP8*- glycans (core 2/LNFP1) interactions. Hydrogen bonding and hydrophobic interactions were shown. C Sequence alignment of VP8* of human RV P genotypes. Sequences of VP8* proteins (residues 81-231) among different human RV P genotypes were analyzed. The alignment was done with DNAMAN. The numbering of the amino acids was based on Rotateq VP8*. The amino acids involved in the glycan binding cavity of P[II] genogroup, P[14], P[11] were colored red, green, blue, respectively. The different residues participating in the glycan binding in human P[6] were highlighted in yellow. Residues 169 and 216 were pointed with red arrows and were colored purple in P[6] and P[19].
The complex structures of the P[4] Indian, P[6] RV3, and P[19] VP8*s with LNFP1 have been determined and superimpositions of these structures showed that these VP8*s shared a common glycan binding site (Fig. 5B, 5C). The non-reducing end (Galβ1-3GlcNAc) of LNFP1 lay in the VP8*s via the same conformation, while Glc at the reducing end showed certain variations. For example, the Glc possesses a conformation closer to the binding cavity of P[6] VP8*, while the same Glc points away from the cavity of other VP8*s (Fig. 5B). The Fuc at the nonreducing end keeps a fixed orientation and was not involved in the direct interaction with the VP8*s. Sequence alignment showed that the amino acids that interact with the glycans, including W81, L167, Y169, G170, G171, R172, W174, T184, T185, R209, E212, and N216, are relatively conserved in the P[8] and P[4] VP8*s, but variations were seen in P[19] and P[6], especially in P[6] VP8*, although its glycan binding site remains at the same location (Fig. 4C). The amino acids of H169, Y170, N171, and S172 in P[6] RV3 VP8* appeared to form stronger interactions with the Glc of LNFP1, bringing the glycan much closer to the binding cavity. Particularly, H169 in P[6] and P[19] VP8*s formed hydrogen bonds with Gal4 of LNFP1 (Fig. 5B), instead, Y169 in the P[4] Indian and P[8] Rotateq VP8*s formed only hydrophobic interactions with Gal4. Residue 216 also appeared to be genotype specific, being N216, N216, V216, T216 in P[8], P[4], P[6], and P[19], respectively, playing various roles. For example, N216 in P[4] and P[8] VP8* contributed a hydrogen bond with the GalII, T216 in P[19] VP8* formed a relatively weak hydrophobic interaction with GalII, while V216 in P[6] VP8* does not involved a direct interaction with LNFP1 (Fig. 5B right panel).
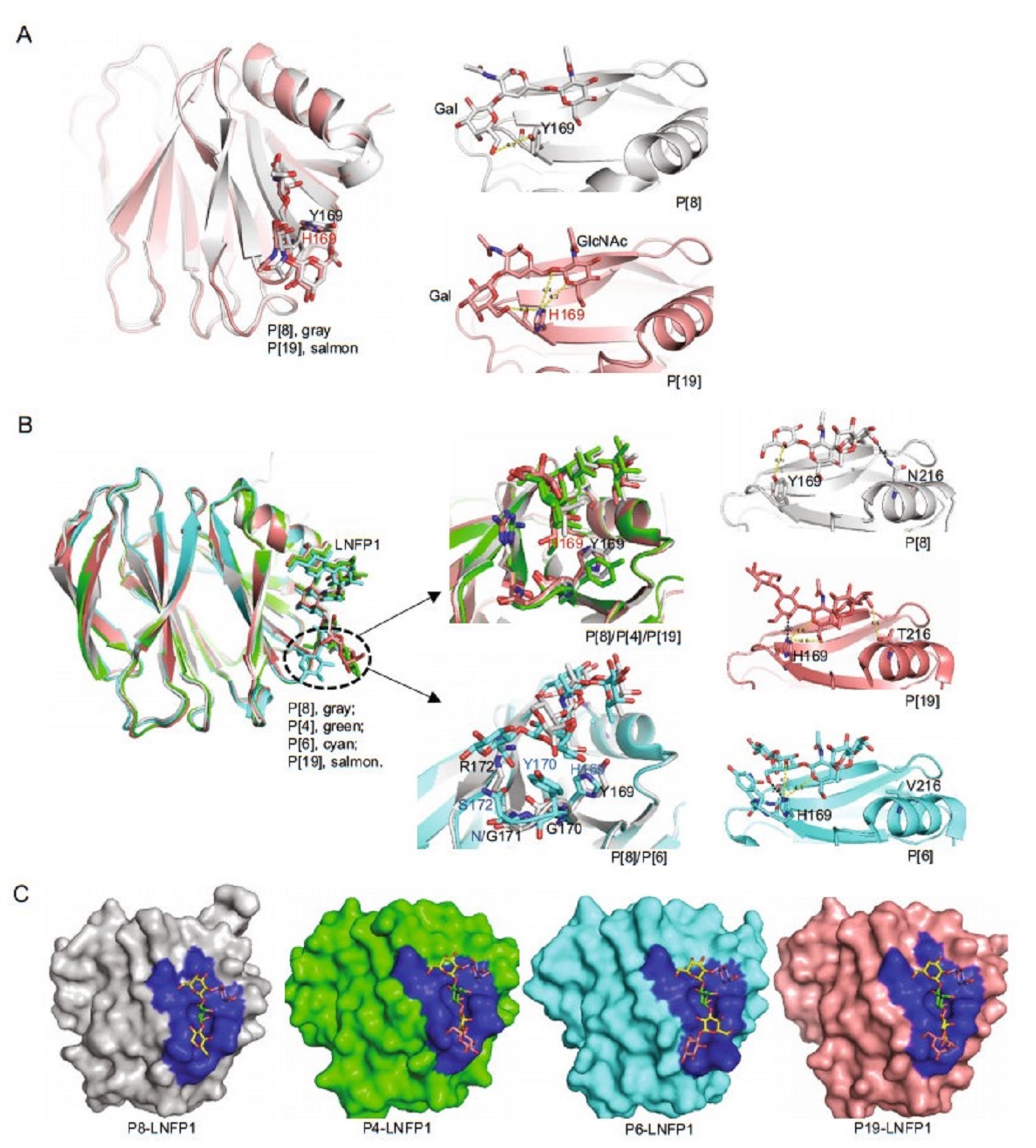
Figure 5. Structural comparison of different P genotype VP8*s-glycan structures. A Structure superimposition of Rotateq P[8] VP8*-core 2 (grey), P[19]-core 2 (salmon) (PDB ID: 5YMS [Ref. (Liu et al. 2017)]), Y169 of P[8] and H169 of P[19] were shown in stick. The hydrophobic interactions were shown in yellow dot lines. B Structure superimposition of Rotateq P[8] VP8*-LNFP1 (grey), P[4]-LNFP1 (green) (PDB ID: 5VX5) [Ref. (Hu et al. 2018)], P[6]-LNFP1 (cyan) (PDB ID: 5VX9) [Ref. (Hu et al. 2018)], P[19]-LNFP1 (salmon) (PDB ID: 5VKS) [Ref. (Liu et al. 2017)]. Residue 169 of P[8]/P[4]/ P[19] and residues 169-172 of P[8] and P[6] were shown in sticks in the middle panel. The interactions between LNFP1 and residue 169/216 in P[8]/P[19]/P[6] VP8* were shown in the right panel. Hydrophobic interactions were shown in yellow dashed lines and hydrogen bond was shown in black dashed line. C Detailed presentation of the LNFP1 binding pocket of P[8]/P[4]/P[6]/P[19] VP8*s. The binding cavity is colored blue in the surface representation. LNFP1 is shown in stick. GlcNAc, green; Gal, yellow; Fuc, slate, Glc, pink.
-
We then tested hemagglutination activity of the P[8] rotavirus VP8*s in form of GST-VP8* fusion proteins to human red blood cells of A, B, and O types. Rotateq P[8]/ P[8]11221075/BELP[8] VP8*s did not hemagglutinate any of the A, B, and O type red blood cells (Fig. 6). The red blood cells of sheep, bovine, pig, and rabbit were also tested but again they were not hemagglutinated by Rotateq P[8] VP8*. Human P[4] and P[6] VP8*s did not show hemagglutination to A, B, O type cells either. As a positive control, the GST tagged P[14] VP8* that recognizes the A type HBGA in the in vitro assays hemagglutinated the A type red blood cells (Fig. 6).
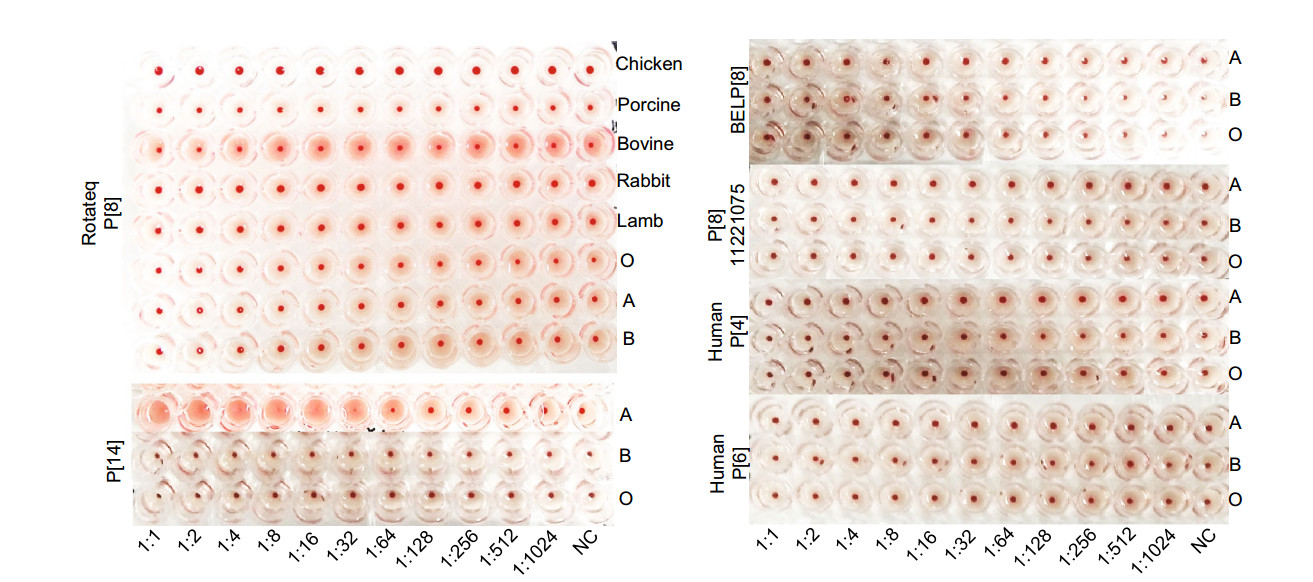
Figure 6. Hemagglutination assay of the recombinant VP8* proteins. GST-VP8* proteins were serially diluted with PBS. Human A, B, O and animal blood cells were applied to the Rotateq P[8] assay. BELP[8], labP[8]-11221075, human P[4]-11151099, P[6]-5311142, P[14] VP8*s were analyzed using human A, B, O red blood cells.
-
Human milk, as well as oligosaccharides LNFP1, core 2, and lactose were used to inhibit P[8] Wa RV infection and replication in cultured MA104 cells (Fig. 7A, 7B). The fluorescent focus was seen as shown in Fig. 7A. P[8] Wa RV infection/replication reduced following incubation of the viruses with human milk at 1:10 (Fig. 7B). Similar blocking effects were observed by LNFP1 glycan at 5 μg/μL. LNFP1 and core 2 glycans at 1 μg/μL also displayed inhibition against P[8] Wa virus infection. Lactose (5 μg/μL) did not show such inhibition. SA11 RV infection was blocked by human milk but not inhibited by LNFP1.
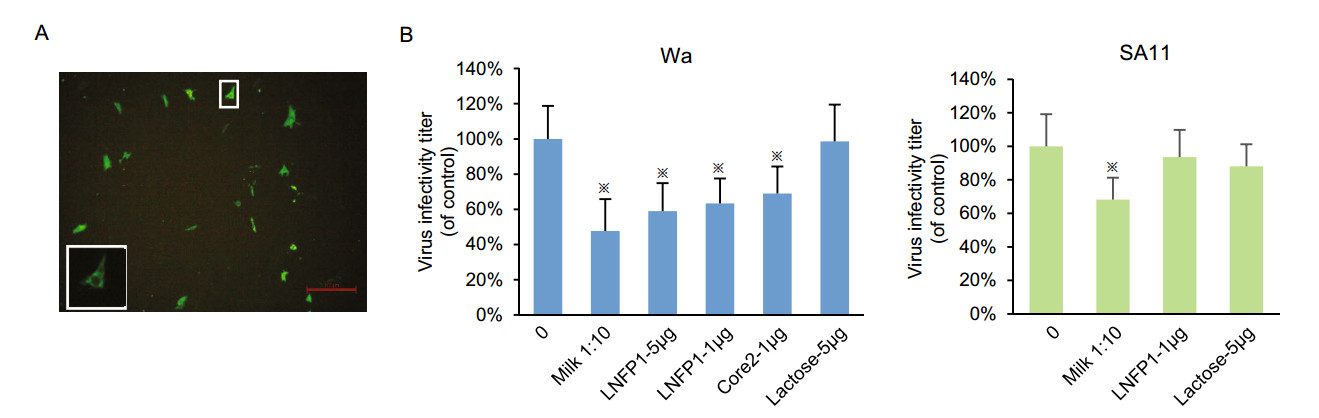
Figure 7. Inhibition of P[8] Wa RV replication in cell culture by human milk and LNFPI/core 2 conjugates. SA11 virus was included as a control. A Representative immunofluorescence microscopy image of MA104 cells infected with P[8] Wa RV. Each green dot represents an infected cell, as shown in the enlarged panel. The red line is a scale bar of 100 μm. The plotted blocking results are shown in panels. B Each bar represents mean % infectivity, with no glycan treatment considered to be 100%. LNFPI, mucin core 2 and lactose oligosaccharides were used at final concentrations of 1 or 5 μg/μL. All assays were carried out twice, with triplicates within each experiment. Error bars represent standard error of the mean. P-values < 0.05 were considered statistically significant [analysis of variance (ANOVA) with Dunnett's correction for multiple comparisons]. The asterisk indicates a statistical difference of P < 0.05.
P[8] RV VP8*s Bound Mucin Core and H Type 1 Glycans
Structural Basis of Rotateq P[8] VP8*-Core 2 Glycan Interaction
Crystal Structure of Rotateq P[8] VP8* Complexed with H-Type 1 Glycan LNFP1
Structural Comparison of the Ligand Binding Sites Among Different RV VP8*s
P[8] VP8*s Did Not Hemagglutinate the ABO Red Blood Cells
Human Milk, LNFP1, and Core 2 Inhibited Cell Infection by P[8] Wa RV
-
Since HBGA glycans were first reported as host attachment factors for RV infection in 2012 (Hu et al. 2012; Huang et al. 2012; Liu et al. 2012), numerous studies have been conducted to explore the complex interactions between the genetically diverse RVs and their polymorphic glycan ligands. These included the phenotypic/functional studies to understand the genotype-specific glycan binding patterns (Huang et al. 2012; Liu et al. 2012; Ramani et al. 2013; Ma et al. 2015; Liu et al. 2016; Jiang et al. 2017), the structural investigations to elucidate the molecular mechanisms behind such interactions (Hu et al. 2012, 2015, 2018; Liu et al. 2017; Sun et al. 2018), and the epidemiology surveillance to associate the human HBGA phenotypes and their susceptibility to RV infection (Imbert-Marcille et al. 2014; Nordgren et al. 2014; Van Trang et al. 2014; Ayouni et al. 2015; Sun et al. 2016b; Barbe et al. 2018). Despite these many studies, our understanding of the globally predominant P[8] RVs in their glycan binding specificity and the mechanism behind virus-glycan binding remain limited. In this study, we verified that P[8] VP8* bound both mucin core 2 glycans and H type 1 antigen via oligosaccharide binding assays. More importantly, we solved the crystal structures of the Rotateq P[8] VP8* proteins complexed with a mucin core 2 trisaccharide and an H type 1 pentasaccharide, respectively, allowing elucidation of the structural basis and mechanism of P[8] RVglycan ligand interactions.
P[8] and P[4] RVs are grouped into the P[II] genogroup together with P[6] and P[19] (Liu et al. 2012). P[4] and P[6] RVs are also predominant worldwide, next to the P[8] genotype and the structural basis of P[4]/P[6]/P[19] VP8* binding to H type 1 glycans have been determined previously, revealing a similar glycan binding site (Liu et al. 2017; Hu et al. 2018; Sun et al. 2018). Here, the crystal structures of P[8] VP8* in complex with core 2 trisaccharide and LNFP1 pentasaccharide verified that the P[8] glycan binding site is indeed highly conserved with the known ones of P[4], P[6], and P[19] VP8*s (Liu et al. 2017; Hu et al. 2018; Sun et al. 2018) in the P[II] genogroup, but not with those of other non-P[II] human RVs (Hu et al. 2012, 2015). We further noted that among the P[II] RVs the amino acids constituting the P[8] glycan binding site are identical to those of the P[4] RVs, while their homology is lower to those of P[19] and even further lower to those of human P[6] glycan binding sites, consistent with their phylogenic relationship (Liu et al. 2012; Hu et al. 2018). These observations may help our understanding on virus-host interaction and prevalence of the globally predominant P[8] RVs.
Our observations that VP8*s of three P[8] RV strains showed obvious binding to mucin core 2, were consistent with those observed previously (Liu et al. 2016). In fact, many tested VP8*s of the P[II] RVs, including human P[4], human P[19], porcine P[19], and porcine P[6] were found to bind mucin core 2 glycan, while human P[6] did not (Liu et al. 2016; Pang et al. 2018; Sun et al. 2018). Mucin core 2 glycans distribute in a variety of cells and tissues (Moran et al. 2011). Our virus inhibition assays showed that core 2 glycan inhibited P[8] RV infection, and similar inhibitory effect was observed to P[19] RV infection (Liu et al. 2017). These data suggested that mucin core glycans, especially core 2, may be important host attachment factors for the P[II] RVs. The structural superimposition and sequence alignment showed the amino acids involved in the interactions with core 2 trisaccharide were relatively conserved in all P[4]/P[8]/P[6]/P[19] RVs except for residue 169. Y169 that is conserved in P[4]/P[8] RVs formed different interactions with core 2 compared with H169 of other P genotypes, which may partly contribute to the differences in binding ability to core 2 glycans. In addition, P[10] VP8* of P[I] genogroup was also reported to recognize the mucin core 2 glycan and shared almost the same amino acids in the glycan binding site (Liu et al. 2016; Pang et al. 2018). Thus, further studies to clarify the specific roles of mucin cores in human RV host ranges and infection are necessary.
Our study confirmed that P[8] VP8* recognize H type 1 antigen (Huang et al. 2012; Ma et al. 2015), but such interaction may be weak. For example, P[8]11221075 VP8* bound conclusively to H type 1 antigens, while the other tested P[8] VP8*s appeared to bind much weaker. P[8] and P[4] VP8*s bound LNT (Galβ1-3GlcNAcb1- 3Galβ1-4Glc) weakly although this glycan contains the H type 1 sequences with one fucose less compared with LNFP1, while P[6] VP8* bound LNT strongly, consistent with a previous study (Liu et al. 2016). Sequence and structural analysis showed that four specific amino acids of P[6] VP8*, spanning from 169 to172 that build part of the glycan binding site, interact strongly with Galβ1-4Glc of LNFP1 at the reducing end, while only relative weak hydrophobic interactions are seen in the P[8]/P[4] VP8*- glycan structures with Glc pointing away from the binding cavity (Fig. 5B). These structural variations explain at least partially the difference of the binding intensity. For the P[19]-LNFP1 complex structure, H169 also formed hydrogen bonds with Gal4 of LNFP1, partially contributing to the relatively strong binding intensity between P[19] and LNFP1/LNT (Liu et al. 2016; Sun et al. 2018). Residue 216 was another amino acid that showed genotype-specific changes and contributed to various interactions in the genotype-related manner as described above, that is, N216 that is specific and relatively conserved in human P[8] and P[4] formed intense interactions with the LNFP1. We speculated that the amino acid variations such as residues 169 and 216 affect the binding intensity to different glycan ligands, which may play a part role in the human RV evolution and prevalence.
Rotateq P[8] RV VP8* bound to core 2 and H type 1 LNFP1 glycans using the same site. The native crystal structure of P[8] Wa was solved previously (Blanchard et al. 2007). Rotateq VP8* structure is close to Wa P[8] VP8* with the RMSD value of 0.277. Interestingly, a glycerol molecule was found in P[8] Wa VP8* at the position equivalent to the GlcNAc of LNFP1 binding site of Rotateq VP8* (Fig. 8A), showing that the P[8] glycan binding site can bind different ligands exquisitely. A recent paper reported the complex structures of P[8]c VP8* with type Ⅰ precursor (LNB, Galβ1-3GlcNAc) and H type 1 (H1, Fuca1-2Galβ1-3GlcNAc) (Rey et al. 2019). We noted that VP8* bound to the glycans via the same site (Fig. 8B). Compared to the structure of P[8] VP8*-LNFP1, LNB and H1 located exactly the position of LNFP1 (Fig. 8B), indicating that P[8] VP8* can accommodate various type Ⅰ related glycans. Previous studies suggested that P[19] VP8*s bind glycans using the GlcNAcb1-3Gal as the basic functional unit (Liu et al. 2017; Sun et al. 2018), which appear the same case in P[8] RV, suggesting that this may be the basic property of all P[II] RVs to bind the GlcNAccontaining glycans. Furthermore, while the GlcNAc-binding pocket was conserved among P[II] RVs, the P[II] RV genotypes exhibited a genotype-specific difference in recognizing the mucin core and type 1-related sequences as shown in our and other studies (Liu et al. 2017; Hu et al. 2018; Sun et al. 2018).
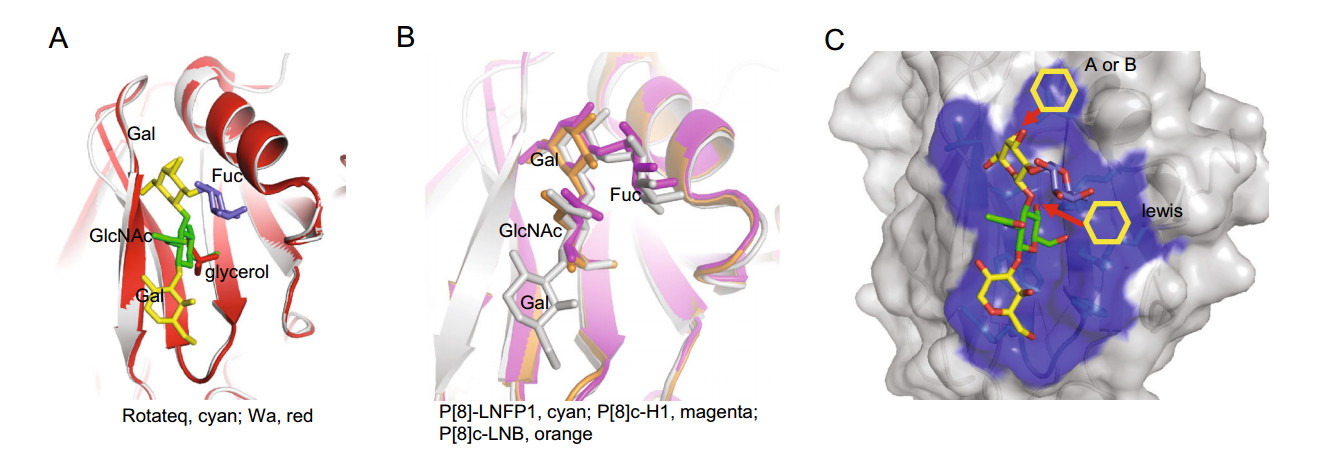
Figure 8. A Structural superimposition of Rotateq P[8] VP8*-LNFP1 (PDB: 6K2O) and Wa P[8] VP8* (PDB: 2DWR). LNFP1 in Rotateq P[8] and the glycerol molecule in Wa P[8] were shown in stick. The LNFP1 was colored as described in Fig. 3. Glycerol was colored red. B Structural comparison of Rotateq P[8] VP8*-LNFP1 (PDB: 6K2O, gray), P[8]c VP8*-H1 (PDB: 6HA0, magenta), P[8]c VP8*-LNB (PDB: 6H9Y, orange). LNFP1, H1, and LNB were presented in stick with color of gray, magenta, orange, respectively. C Surface presentation of LNFP1 binding site in Rotateq P[8] VP8* and the supposed elongation of A/B/lewis antigens.
The type 1 precursor Galβ1-3GlcNAc could develop into different H, A, B, and Lewis HBGAs by adding one specific saccharide step by step. Interestingly, P[8] RV VP8* did not interact with A/B/Lewis type HBGAs as shown by both oligosaccharide binding and red blood cell hemagglutination assays. The binding of P[8] VP8*s to saliva samples was independent on the ABO types (Sun et al. 2016a). In addition, two previous epidemiological studies found that P[8] RV infect preferably secretors irrespective of the individual's ABO types (Van Trang et al. 2014; Sun et al. 2016c), while another recent study demonstrated that in vitro infection of transformed cell lines by P[8] RVs was independent of HBGAs expression (Barbe et al. 2018). These results supported the notion that P[8] RV VP8* may not recognize the ABO HBGAs. Structural analysis showed that a linkage of a GalNAc (A) or a Gal (B) to the O3 of GalII is sterically possible, but an addition of fucose to GlcNAc by α1-4 linkage to generate the Lewis antigens would cause steric hindrance (Fig. 8C). This may partly contribute to the fact that P[8] RV VP8* did not show obvious binding to a Lewis antigen in the oligosaccharide assay. Another possibility is that the binding to ABO HBGAs by P[8] VP8* may be too weak to be detected by an ELISA assay. A recent study using NMR titration experiments and NMR derived high ambiguity driven docking (HADDOCK) methods demonstrated that P[8] VP8* bound the Lewis b antigen with a Kd value of 43.4 mM via an undescribed pocket formed by two bsheets (Xu et al. 2019). Further studies to clarify these issues are expected.
The available data indicate that all P[8], P[4], P[6], and P[19] interact with type Ⅰ HBGA pentasaccharide LNFP1 that is a main component of the human milk oligosaccharides (HMOs) (Kunz et al. 2000). HMOs are believed to contain prebiotic or anti-microbial effector molecules benefiting neonates (Etzold and Bode 2014; Morozov et al. 2018). Accordingly, the VP8*s of human neonatal RVs (P[11]) bound HMOs independent from sialic acids, exhibiting a unique glycan specifically (Yu et al. 2014). Here, we showed that human P[8] VP8* recognized the HMO LNFP1 via the same binding mode as that of P[4]/ P[6]/P[19] VP8*s. We and others (Liu et al. 2017; Hu et al. 2018) further showed that LNFP1 glycan inhibited infections of human P[8]/P[4]/P[6]/P[19] RVs infection, suggesting that LNFP1 glycans in the human milk may serve as decoy ligands to block the infectivity of human RVs, including P[8]/P[4]/P[6]. This may partly contribute to the reduced risk of breast-fed infants to acquire RV infection. However, a recent report showed that HMOs are not decoy receptors for G10P[11] and specific HMOs enhanced the infectivity of the neonatal-derived RV vaccine (P[11]), while HMOs had no significant effect on the infectivity of the globally dominant P[4] and P[8] human RVs (Ramani et al. 2018). These data discrepancies indicate the RVHMOs interactions are complex and need further studies for clarification.
In summary, we have redefined the glycan binding specificity of the globally predominant P[8] RVs to verify the mucin core 2 as well as the H type Ⅰ antigen as host ligands. Compared to that of Hu et al. (2018) which reported the P[4]/P[6] VP8*-LNFP1 structures, we determined the structure of P[8] VP8*-LNFP1, verifying the glycan binding site and interaction mechanism for P[8] RVs. For Rey et al. (2019) that determined P[8] VP8* in complex with H type 1 antigens, we demonstrated that P[8] VP8* not only interact with H type 1 antigens, but also mucin core antigens based on crystal structures of VP8*- LNFP1 and VP8*-core 2. Our study elucidated the structural basis of P[8] VP8* accommodating different glycan ligands and provided phenotypic and structural information to comprehensively understand the virus-host interaction and epidemiology of the globally prevalent P[8].
-
We are grateful to Jingyu Yan in Dalian Institute of Chemical Physics for kindly providing the free LNFP1 oligosaccharide purified from human milk. We would like to thank George F. Gao, Jinhua Yan, and Yi Shi in the Institute of Microbiology for the help in data analysis. We thank Huiying Li, Xiaohui Gao, Qiuyu Huang, Yu Qing, Lili Pang, Miao Jin, Xiangyu Kong in our lab for their help in the experiments. We appreciate the help of Yuan Yuan, Min Zhao, Yan Li, Xu Yang, Ruchao Peng, and Han Wang in the Institute of Microbiology in X-ray data collection. The assistance by the staff at the Shanghai Synchrotron Radiation Facility (SSRFbeamline 17U) is acknowledged. This research was supported by grants from the National Science and Technology Major Project (2018ZX10711-001) and the National Natural Science Foundation of China (NSFC) (No. 81601813).
-
ZD, XS, and DL designed the experiments. XS, LD, DL, and MW performed the protein expression and glycan binding experiments. XS, LD, QZ, and HW performed the virus inhibition assay. JQ solved the structures. XS and JQ analyzed the structural data. XS, RB, WC interpreted the glycan binding data. XS wrote the draft manuscript. ZD, XS, and MT revised the manuscript.
-
The authors declare that they have no conflict of interest.
-
The study was approved by the Medical Ethics Committee at National Institute for Viral Disease Control and Prevention, China CDC, Beijing, China and by the Institutional Animal Care and Use Committee (IACUC) of National Institute for Viral Disease Control and Prevention, China CDC. All the experimental procedures were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People's Republic of China.














 DownLoad:
DownLoad: