HTML
-
Coronaviruses are enveloped viruses of the order Nidovirales harboring a single-stranded positive sense RNA genome. With approximately 31 kb, their genomes are the largest among RNA viruses (Masters and Perlman 2013; Siddell and Snijder 2005). Coronaviruses are genetically classified into four major genera: Alpha-, Beta-, Gamma-, and Deltacoronaviruses (Li 2016). While the first two primarily infect mammals, the two latter predominantly infect birds (Tang et al. 2015). According to the typical severity of the diseases, physicians categorize coronaviruses infecting humans into two types: those accompanied with rather mild pathogenicity for healthy adults such as the causative agents of common cold (e.g., HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU) as opposed to those viruses frequently causing serious and life-threatening diseases (e.g., SARS-CoV and MERS-CoV) (Kuiken et al. 2003; Kuypers et al. 2007; Peiris et al. 2003; Yin and Wunderink 2018).
In December 2019, a novel coronavirus was first recognized in Wuhan, China. The virus first spread to other Chinese provinces and later throughout the entire world. To date, no definitive patient zero has been identified and there is an ongoing debate on when and where the first animal-to-human transmission may have occurred. The virus was initially referred to as 2019 novel coronavirus (2019-nCoV) by the World Health Organization (WHO) on January 12, 2020 (Chen Yu et al. 2020). Later, it was renamed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses (ICTV) on February 11, 2020 (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses 2020). The new name caused great concern and controversy in the field. More than 20 renowned virologist in China—including several individuals who either contributed to the discovery of the new coronavirus or established therapies for new coronavirus-induced pneumonia—expressed their concerns and instead proposed the name Human coronavirus 2019 (HCoV-19) (Jiang et al. 2020; Zhang D et al. 2020). The disease, caused by SARS-CoV-2, was named Coronavirus disease 2019 (COVID-19) on February 12, 2020 and was declared a pandemic on March 11, 2020 by WHO (World Health Organization 2020b). We summarize the events mentioned above in Table 1.
Time Event Organization 2020.01.12 2019 novel coronavirus (2019-nCoV) WHO 2020.02.11 Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) ICTV 2020.02.12 Coronavirus disease 2019 (COVID-19) WHO 2020.03.11 A pandemic WHO Table 1. Related events of novel coronavirus.
As of September 7, 2020, there have been 26, 994, 442 confirmed cases of COVID-19, including 880, 994 deaths worldwide (https://covid19.who.int/). To mitigate the impact on human health, rapid investigation of the etiology of SARS-CoV-2 is warranted. Here, we review our current understanding of the genome organization, the virion morphology, the receptor usage, and origin of SARS-CoV-2.
-
By pairwise protein sequence analysis, the seven conserved replicase domains in ORF1ab used for CoV species classification, show 94.6% amino acid sequence identity of SARS-CoV-2 with SARS-CoV. At first glance, this implied that the two viruses are so closely related that they could even be assigned to the same species (Zhou P et al. 2020). However, more detailed phylogenetic analyses, which took the complete genomes into account, uncovered that SARS-CoV-2 is distinct from SARS-CoV and MERS-CoV. Moreover, SARS-CoV-2, together with two bat-derived SARS-like strains, ZC45 and ZXC21, form a distinct clade in the lineage B of the subgenus Sarbecovirus. Because the sequence identity in conserved replicase domains (ORF 1ab) between SARS-CoV-2 and other members of the Betacoronavirus genus is less than 90%, SARS-CoV-2 represents a novel betacoronavirus belonging to the Sarbecovirus subgenus of the Coronaviridae family (Lu et al. 2020; Zhu et al. 2020).
To understand the origin of the SARS-CoV-2 and its genetic relationship with other coronaviruses, Xintian Xu et al. performed a phylogenetic analysis on a collection of coronavirus sequences from various sequence depositories. The result showed that SARS-CoV-2 viruses cluster together in the phylogenetic tree of CoVs and belong to the Betacoronavirus genus (Xu et al. 2020). Although SARS-CoV-2 exhibits striking similarities to some betacoronaviruses detected in bats, it is distinct from known bat coronaviruses, SARS-CoV (Chan et al. 2020; Lu et al. 2020; Malik et al. 2020; Zhou P et al. 2020; Zhu et al. 2020), and MERS-CoV (Lu et al. 2020; Zhu et al. 2020). Additionally, SARS-CoV-2 shares higher sequence homology to the genome of SARS-CoV than to that of MERS-CoV (Xu et al. 2020). Notably, further studies have shown that SARS-CoV-2 is closely related to bat SARS-like CoVs such as bat-SL-CoVZC45 (Lu et al. 2020; Wu F et al. 2020; Zhu et al. 2020) and bat-SL-CoVZXC21 (Chan et al. 2020; Lu et al. 2020; Malik et al. 2020; Zhu et al. 2020). Meanwhile, even closer homologs have been identified: Carmine Ceraolo and Federico M. Giorgi found that the Bat-CoV RaTG13 genome (Gisaid EPI_ISL_402131) has 96.2% sequence identity to the SARS-CoV-2 reference sequence (NC_045512.2) (Ceraolo and Giorgi 2020). In addition, other reports have investigated coronavirus of non-bat origin, such as Pangolin-CoV (Chen Yun et al. 2020a; Liu P et al. 2020; Xiao et al. 2020; Zhang LS et al. 2020; Zhang T et al. 2020). We have compiled the specific nucleotide sequence identity between SARS-CoV-2 and other coronaviruses in Table 2. Based on these similarities to bat and pangolin CoVs, it is tempting to speculate that a merger of viruses derived from these two nocturnal animals may have preceded the animal-to-human transmission event.
Nucleotide sequence identity (%) Other coronaviruses Accession number References 96.2 Bat-RaTG13-CoV Not available Ceraolo and Giorgi 2020 89 Bat-SL-CoVZXC21 MG772934 Chan et al. (2020) 82 Human SARS-CoV BJ01 AY278488 Chan et al. (2020) 82 Human SARS-CoV Tor2 AY274119 Chan et al. (2020) 87.99 Bat-SL-CoVZC45 MG772933 Chen Yan et al. (2020) 80 SARS-CoV Not available Chen Yan et al. (2020) 90.23 Pangolin-CoV-2020 GWHABKW00000000 Liu P et al. (2020) 87.99 Bat-SL-CoVZC45 MG772933 Lu et al. (2020) 87.23 Bat-SL-CoVZXC21 MG772934 Lu et al. (2020) ~ 79 SARS-CoV Not available Lu et al. (2020) ~ 50 MERS-CoV Not available Lu et al. (2020) ≥ 88.2 Bat-SL-CoVZC45 MG772933 Malik et al. (2020) ≥ 88.2 Bat-SL-CoVZXC21 MG772934 Malik et al. (2020) 89.1 Bat-SL-CoVZC45 MG772933 Wu F et al. (2020) 80–98 Pangolin-CoV Not available Xiao et al. (2020) 91.02 Pangolin-CoV Not available Zhang T et al. (2020) 79.6 SARS-CoV BJ01 AY278488 Zhou P et al. (2020) > 85 Bat-SL-CoVZC45 MG772933 Zhu et al. (2020) Table 2. Nucleotide sequence identity between SARS-CoV-2 and other coronaviruses.
-
The average evolutionary rate for coronaviruses is approximately 10−4 nucleotide substitutions per site per year, with mutations arising during every replication cycle (Su et al. 2016). Therefore, it is striking that the sequences of SARS-CoV-2 from different patients are almost identical, with greater than 99.9% sequence identity (Ceraolo and Giorgi 2020; Lu et al. 2020; Xu et al. 2020; Zhou P et al. 2020). Despite the RNA proofreading capacities of coronaviruses (Romano et al. 2020), this finding suggests that SARS-CoV-2 originated from a single transmission event and was identified relatively rapidly thereafter, prior to the onset of relevant speciation processes.
Although sequence diversification between strains of SARS-CoV-2 is rather small and it is difficult to separate them in phylogenetic cladograms, isolates of SARS-CoV-2 can still be divided into 6 genotypes, indicating that different mutations occurred in different patients (Zhang LS et al. 2020). Xialu Tang et al. analyzed SARS-CoV-2 virus strains and found that 101 showed complete linkage between the two single nucleotide polymorphisms: 72 strains exhibited a "CT" haplotype (defined as the "L" type), and 29 strains exhibited a "TC" haplotype (defined as the "S" type) (Tang et al. 2020). In the study of Rozhgar A. Khailany et al., 116 unique variants were identified, comprising 46 missense, 52 synonymous, 2 insertions, 1 deletion, and 14 noncoding alleles. The most common variants were C8782T (affecting the ORF1ab gene), T28144C (affecting the ORF8 gene), and C29095T (affecting the N gene) (Khailany et al. 2020). Syed Mohammad Lokman et al. investigated genomes of SARS-CoV-2 and found variations in 483 positions, resulting in 130 synonymous and 228 nonsynonymous variants (Lokman et al. 2020). Additionally, Jun-Sub Kim et al. identified 767 synonymous and 1352 nonsynonymous mutations in the SARS-CoV-2 genome (Kim et al. 2020). Importantly, a novel isolate of the SARS-CoV-2 virus carrying a point mutation in the Spike protein (D614G) has emerged and rapidly surpassed others in prevalence (Korber et al. 2020). Recent studies provide functional evidences that the D614G mutation in the Spike protein increases transduction of the virus in human cells. Zharko Daniloski et al. showed that in multiple cell lines, SARS-CoV-2-pseudotyped lentiviral particles carrying the D614G mutation was up to eightfold more effective at transducing cells than wild-type; G614 Spike was more resistant to proteolytic cleavage, which may suggest a possible mechanism for the increased transduction (Daniloski et al. 2020). Lizhou Zhang et al. observed pseudoviruses-G614 infected hACE2-293T cells with approximately ninefold higher efficiency than did pseudoviruses-D614, and confirmed that the D614G mutation enhances virus infection through reducing S1 shedding and increasing total S protein incorporated into the virion (Zhang LZ et al. 2020). Moreover, in the study of Jie Hu et al., about 7% convalescent sera showed decreased neutralizing activity against pseudoviruses-G614, indicating that D614G mutation changed the antigenicity of S protein, thereby decreasing neutralization sensitivity to individual convalescent sera (Hu et al. 2020). However, another study demonstrated controversy results showing that the G614 variant was sensitive to neutralization by polyclonal convalescent sera (Korber et al. 2020).
In the light of discussions concerning the occurrence of mutations, which apparently alter the phenotype of SARS-CoV-2 such as the D614G spike mutation, more attention should be given to the prevalence of genetic variations and their potential biological consequences. If such variations affect T cell epitopes and/or antibody epitopes, such surveillance would be highly relevant in terms of vaccine development and the establishment of collective 'herd" immunity.
-
Recombination often occurs between coronaviruses (Su et al. 2016). Although similarity plots suggest a possible recombination event between SARS-CoV-2 and SARS-CoV or SARS-like CoVs, no clear evidence of recombination exists between these genomes (Wu F et al. 2020). Roujian Lu et al. did phylogenetic analyses of the major encoding regions of representative members of the subgenus Sarbecovirus, and found that potential recombination events in the bat coronaviruses rather than SARS-CoV-2 (Lu et al. 2020). Therefore, recombination may not explain the emergence of the virus, although this inference might change if more closely related animal viruses are identified.
Arinjay Banerjee et al. discussed a high recombination potential in the ORF1ab gene region of SARS-CoV-2 and MERS-CoV, with three homologous genomic regions that could promote recombination. For such recombination events, co-infections must occur in common host cells. The SARS-CoV-2 receptor angiotensin-converting enzyme 2 (ACE2) and MERS-CoV receptor dipeptidyl peptidase 4 (DPP4) are co-expressed at high levels in the human small intestine and other tissues, stimulating the authors to speculate whether recombination events between the novel coronavirus and MERS-CoV may be possible in the future (Banerjee et al. 2020).
Emergence of a Novel Human Pathogenic Betacoronavirus
Genomic Variation
Gene Recombination
-
According to transmission electron microscopy (EM) and cryo EM, the morphology of the SARS-CoV-2 particle is round, oval or pleomorphic, with a diameter of 60–140 nm. Virions have distinct spikes, protruding approximately 9 to 12 nm from the membrane surface that give virions the appearance of a solar corona or medieval crown and hence gave this virus family its name (Lu et al. 2020; Zhou P et al. 2020; Zhu et al. 2020). The composition of envelope of coronaviruses is reminiscent to the membrane of the former host cell, comprising membrane proteins and lipid bilayers. Steffen Klein et al. measured the lipid bilayer separation of SARS-CoV-2 virions: a phospholipid monolayer separation was 3.6 nm whereas the plasma membrane was 3.9 nm (Klein et al. 2020). The structure of the nucleocapsid is a symmetrical helix, formed by the combination of positive-strand RNA and nucleocapsid protein (N) (Peiris et al. 2004).
The single-stranded RNA genome of the SARS-CoV-2 reference sequence is 29, 891 nucleotides in size and encodes 9860 amino acids. The G + C content is 38% (Chan et al. 2020). SARS-CoV-2 shows a genome organization typical for betacoronaviruses: a 5′ untranslated region (5′-UTR), an RNA polymerase open reading frame (ORF1ab) followed by genes encoding for the spike (S) protein, the envelope (E) protein, the membrane (M) protein, the nucleocapsid (N) protein, a 3′ untranslated region (3′-UTR), and several nonstructural open reading frames (Chan et al. 2020; Wu F et al. 2020; Zhu et al. 2020). The genome gives rise to at least 12 canonical proteins and 23 non-canonical translation products (Finkel et al. 2020).
According to GenBank, the protein lengths of these five important proteins are 7096 aa (ORF1ab), 1273 aa (S protein), 75 aa (E protein), 222 aa (M protein) and 419 aa (N protein). The RNA polymerase gene occupies 67% of the coronavirus genome, while all other structural proteins and accessory proteins are encoded by the remainder of the genome (Chen Yu et al. 2020; Guan et al. 2003; Wu A et al. 2020). One study predicted putative functions and proteolytic cleavage sites of ORF1ab, including 16 nonstructural proteins, by bioinformatics (Chan et al. 2020). The S protein, integrated over the surface of the virus, mediates the attachment of the virion to the host cell surface receptors and enforces the fusion between viral and host cell membranes, facilitating viral entry into the host cell; the E-protein plays important roles in virus invasion and reproduction, including virus assembly, membrane permeability of the host cell and the virus–host cell interaction; the M-protein is considered the central organizer for coronavirus assembly, which is most abundant in the viral surface; the N protein, coating the viral RNA genome, plays a vital role in its replication and transcription (Boopathi et al. 2020). The protein length, location, and function of these four structural proteins are summarized in Table 3. At present, the conservation of the gene regions encoding ORF1ab (especially the RNA-dependent RNA polymerase [RdRp]), the E, and N protein have been selected to develop quantitative Reverse-Transcriptase-PCR (qRT-PCR)-based nucleic acid detection methods (Chu et al. 2020; Corman et al. 2020; Lu et al. 2020; Wu F et al. 2020; Zhou P et al. 2020). Based on its abundance, methods recognizing viral proteins such as mass-spectrometry and in-cell-ELISAs frequently target the N protein (Ihling et al. 2020; Nikolaev et al. 2020; Schöler et al. 2020).
Structural protein Protein length (aa) Location Function ( Boopathi et al. 2020 )S protein 1273 21, 563..25, 384 Receptor binding and membrane fusion E protein 75 26, 245..26, 472 Virus invasion and reproduction M protein 222 26, 523..27, 191 Virus assembly N protein 419 28, 274..29, 533 Replication and transcription Table 3. Protein length, location and function of S protein, E protein, M protein and N protein of SARS-CoV-2.
-
The spike glycoprotein forms spikes on the surface of coronaviruses, attaching the viruses to host cell receptors and catalyzing virus–cell membrane fusion. For this purpose, the receptor-binding domain (RBD) in the spike glycoprotein directly binds receptors on the surface of host cells. The amino acid sequences and predicted protein structure are highly similar between SARS-CoV-2 and SARS-CoV RBD, suggested that SARS-CoV-2 may also use human angiotensin-converting enzyme 2 (ACE2) as a cellular entry receptor, facilitating human-to-human transmission (Letko et al. 2020; Lu et al. 2020; Wan et al. 2020; Wu F et al. 2020; Zhu et al. 2020).
To determine whether SARS-CoV-2 uses the ACE2 as receptor, virus infectivity studies were conducted, showing that SARS-CoV-2 can use all but mouse ACE2 as entry receptors (Zhou P et al. 2020). Additional reports revealed a unique structural feature of the RBD of SARS-CoV-2—a unique phenylalanine F486 that confers potentially higher affinity binding for its receptor than found with absent in SARS-CoV (Chen Yun et al. 2020; Wrapp et al. 2020). The major hot spot amino acids involved in the binding identified by interaction analysis after simulations include Lys 31, His 34, Glu 35, Glu 37, Asp 38, and Tyr 83 residues of the ACE2 receptor and Lys 417, Asn 487, Gln 493, Gln 498, and Tyr 505 residues in the SARS-CoV-2 S-protein RBD (Veeramachaneni et al. 2020). The distribution of ACE2 in human tissues and organs using The Human Protein Atlas (https://www.proteinatlas.org/ENSG00000130234-ACE2/tissue) reveals high ACE2 expression in the duodenum, small intestine, gallbladder, kidney and testis (Fig. 1)—which may explain the multiorgan tropism of SARS-CoV-2. Interestingly, recent studies demonstrate that ACE2 is expressed at very low levels—if at all—in the lung and the upper respiratory tract, which indicates that SARS-CoV-2 may have alternative receptors (Zhang Z et al. 2020; Hikmet et al. 2020).
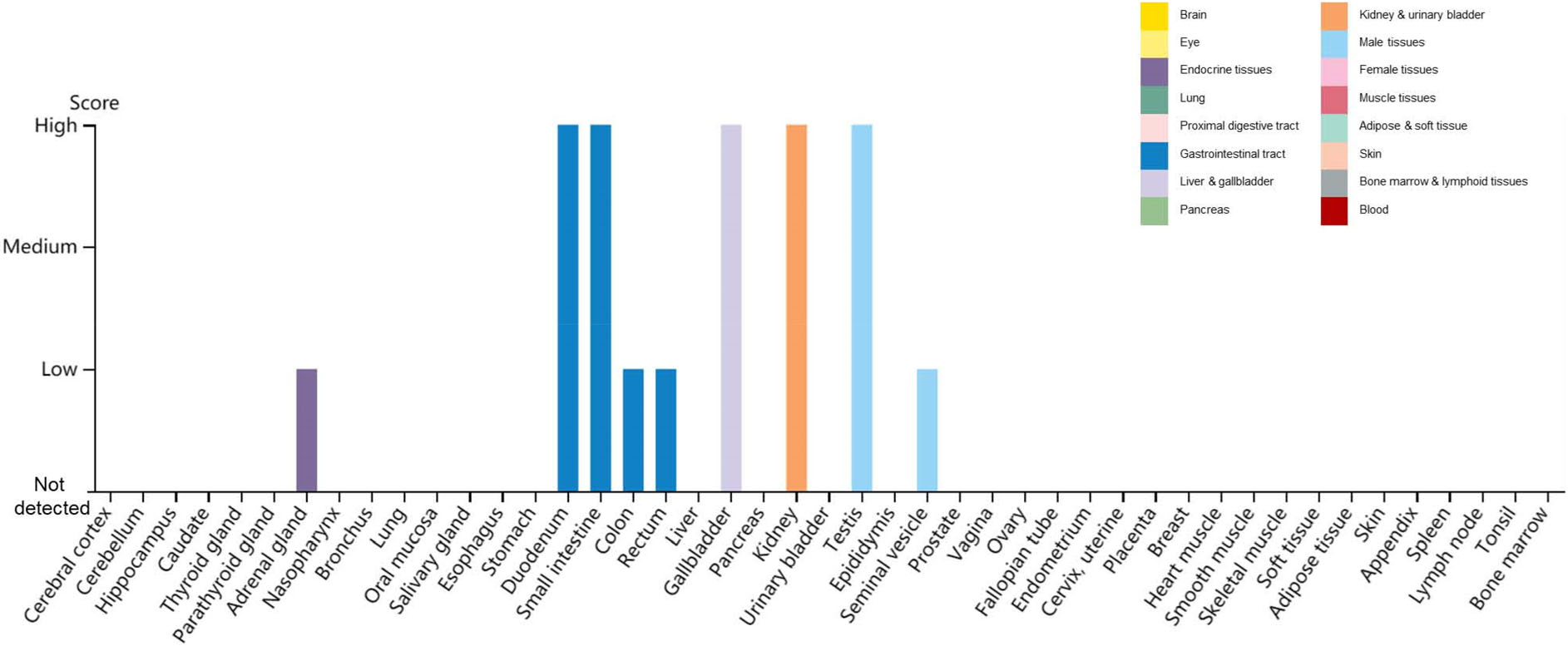
Figure 1. Distribution of ACE2 in human tissues and organs. The expression levels of ACE2 protein in the annotated tissues and organs are presented based on the immunohistochemistry data provided by the Human Protein Atlas (HPA). Color-coding is based on tissue groups, each consisting of tissues with functional features in common. Protein expression score is based on immunohistochemical data manually scored with regard to staining intensity (negative, weak, moderate or strong) and fraction of stained cells (< 25%, 25–75% or > 75%). Each combination of intensity and fractions is automatically converted into a protein expression level score as follows: negative—not detected; weak combined with < 25%—not detected; weak combined with either 25–75% or 75%—low; moderate combined with < 25%—low; moderate combined with either 25–75% or 75%—medium; strong combined with < 25%—medium; strong combined with either 25–75% or 75%—high.
Another membrane protein essential for SARS-CoV-2 entry into cells is the transmembrane serine protease 2 (TMPRSS2), which cleaves the Spike protein triggering its fusogenic activity and enabling the entry into host cells. TMPRSS2 is essential for SARS-CoV-2 entry into cells—at least in absence of exogenously added proteases such as trypsin. Accordingly, the pharmacologic inhibition of TMPRSS2 by protease inhibitor such as camostat mesylate inhibits the replication of SARS-CoV-2 (Hoffmann et al. 2020). Two natural compounds, Wi-A and Wi-N, bind and stably interact with the catalytic site of TMPRSS2, showing the potential to block TMPRSS2 (Kumar et al. 2020). Additionally, TMPRSS2 is downregulated by betacoronavirus infection (Bahir et al. 2009). The distribution of TMPRSS2 in human tissues and organs using The Human Protein Atlas (https://www.proteinatlas.org/ENSG00000184012-TMPRSS2/tissue) reveals the highest expression of TMPRSS2 in the kidney, followed by the parathyroid gland, stomach, pancreas, epididymis and prostate (Fig. 2).
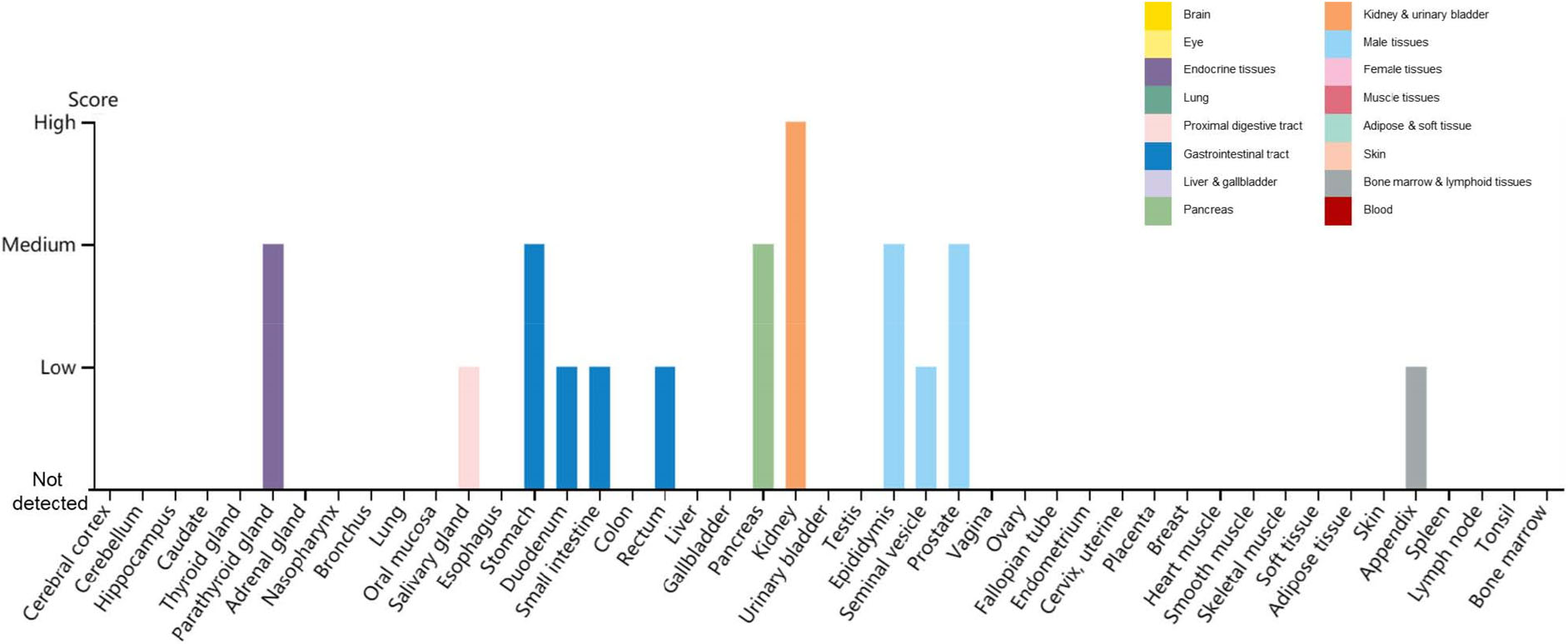
Figure 2. Distribution of TMPRSS2 in human tissues and organs. The expression levels of TMPRSS2 protein in the annotated tissues and organs are presented based on the immunohistochemistry data provided by the Human Protein Atlas (HPA). Color-coding is based on tissue groups, each consisting of tissues with functional features in common. Protein expression score is based on immunohistochemical data manually scored with regard to staining intensity (negative, weak, moderate or strong) and fraction of stained cells (< 25%, 25–75% or > 75%). Each combination of intensity and fractions is automatically converted into a protein expression level score as follows: negative—not detected; weak combined with < 25%—not detected; weak combined with either 25–75% or 75%—low; moderate combined with < 25%—low; moderate combined with either 25–75% or 75%—medium; strong combined with < 25%—medium; strong combined with either 25–75% or 75%—high.
Recently reported, a protein, Neuropilin-1 (NRP1), is important for the invasion of SARS-CoV-2 into host cells. To infect cells, the S protein of SARS-CoV-2 must be cleaved to two associated polypeptides: S1 and S2 (Wrapp et al. 2020). There is a polybasic Arg–Arg–Ala–Arg (RRAR) C-terminal sequence on S1, which means such C-terminal sequence conforming to the 'C-end rule' (CendR) are known to bind to and activate neuropilin receptors (NRP1 and NRP2) at the cell surface (Pang et al. 2014; Teesalu et al. 2009). James L. Daly et al. demonstrated that, in addition to engaging ACE2, S1 can bind to NRP1 through the canonical CendR mechanism, by using immunoprecipitation, site-specific mutagenesis, structural modelling, and antibody blockade (Daly et al. 2020). In the research of Ludovico Cantuti-Castelvetri et al., they also found that the cellular receptor NRP1 significantly potentiates SARS-CoV-2 infectivity (Cantuti-Castelvetri et al. 2020). The distribution of NRP1 in human tissues and organs using The Human Protein Atlas (https://www.proteinatlas.org/ENSG00000099250-NRP1/tissue) reveals that NRP1 is expressed in many tissues and organs (Fig. 3). Additionally, it has been shown that SARS-CoV-2 does not use other coronavirus receptors, such as aminopeptidase N and dipeptidyl peptidase 4 (Zhou P et al. 2020).
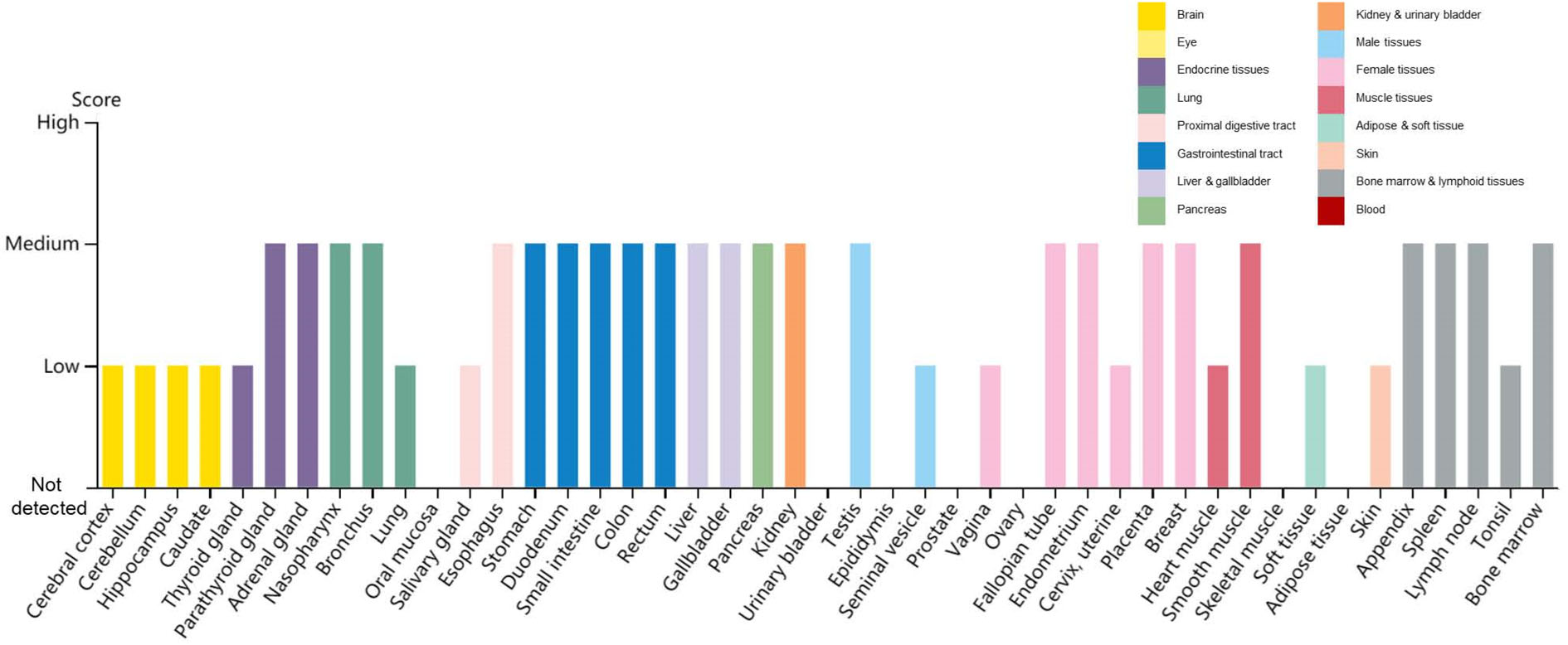
Figure 3. Distribution of NRP1 in human tissues and organs. The expression levels of NRP1 protein in the annotated tissues and organs are presented based on the immunohistochemistry data provided by the Human Protein Atlas (HPA). Color-coding is based on tissue groups, each consisting of tissues with functional features in common. Protein expression score is based on immunohistochemical data manually scored with regard to staining intensity (negative, weak, moderate or strong) and fraction of stained cells (< 25%, 25–75% or > 75%). Each combination of intensity and fractions is automatically converted into a protein expression level score as follows: negative–not detected; weak combined with < 25%—not detected; weak combined with either 25–75% or 75%—low; moderate combined with < 25%—low; moderate combined with either 25–75% or 75%—medium; strong combined with < 25%—medium; strong combined with either 25–75% or 75%—high.
-
Since the outbreaks of SARS in 2002 and MERS in 2012, the possibility of the transmission of coronaviruses from animals to humans has been proven (Cauchemez et al. 2013; Cui et al. 2019). There is high sequence similarity (> 99%) between all sequenced SARS-CoV-2 genomes available, and the nearest most closely related bat coronavirus sequence has 96.2% sequence identity, confirming the possibility of arguing for a zoonotic origin of SARS-CoV-2 (Ceraolo and Giorgi 2020). According to social media reports, in addition to seafood, the South China Seafood Mark et also sells snakes, birds and other small mammals. WHO reported that environmental samples taken from the marketplace were positive for the novel coronavirus (SARS-CoV-2), but no specific animal association has been identified (World Health Organization 2020a). Many scientists have researched potential hosts of SARS-CoV-2. We have summarized the origin permissiveness and potential origins of SARS-CoV-2 in Table 4.
Origin Support Opposition Bat 1. SARS-CoV-2 has 96.2% homology with bat coronavirus (RTG13) (Zhou P et al. 2020)
2. SARS-CoV-2 is most closely related to two SARS-like coronavirus sequences isolated from bats (Chan et al. 2020; Malik et al. 2020; Zhang LS et al. 2020)
3. SARS-CoV-2 has 100% amino acid similarity to BAT-SL-CoVZC45 in nsp7 and E proteins (Wu F et al. 2020)
4. SARS-CoV-2 most likely originated from a bat SARS-like coronavirus by Bayesian phylogeographic reconstruction (Benvenuto et al. 2020)
5. SARS-CoV-2 can infect bat intestinal cells (Zhou J et al. 2020)Unreported Pangolin 1. Pangolin coronavirus genomes have more than 85.5% similarity to SARS-CoV-2 (Lam et al. 2020; Xiao et al. 2020; Zhang et al. 2020f)
2. The amino acid identities of pangolin coronavirus and SARS-CoV-2 in the E, M, N and S genes were 100%, 98.2%, 96.7% and 90.4%, respectively (Xiao et al. 2020)1. Phylogenetic analyses do not support that SARS-CoV-2 arose directly from Pangolin-CoV (Liu P et al. 2020) Snake 1. SARS-CoV-2 has the highest similarity with the synonymous codon usage bias of snakes (Zhou P et al. 2020) 1. The study has several limitations (Zhang C et al. 2020)
2. The ACE2 of snake lost the capability to associate with S protein (Luan et al. 2020)Turtle 1. The interaction between the key amino acids of S protein RBD and ACE2 indicates that turtles may be a potential intermediate host for transmitting SARS-CoV-2 to humans (Liu Z et al. 2020) 1. The ACE2 of turtle lost the capability to associate with S protein (Luan et al. 2020) Mink 1. By comparing the infectivity patterns of all viruses hosted on vertebrates, mink viruses show a closer infectivity pattern to SARS-CoV-2 (Guo et al. 2020)
2. Minks turned out to be highly permissive for SARS-CoV-2 (Enserink 2020)
3. Minks were infected by SARS-CoV-2 (Oreshkova et al. 2020)Unreported Ferret 1. SARS-CoV-2 can replicate in the upper respiratory tract of ferrets (Shi et al. 2020) Unreported Cat 1. Cats are highly susceptible to SARS-CoV-2 (Shi et al. 2020)
2. Cats acquire SARS-CoV-2 infection under natural conditions (Zhang Q et al. 2020)
3. Infected cats can spread SARS-CoV-2 from one cat to another (Halfmann et al. 2020)Unreported Dog 1. Two dogs in Hong Kong are infected with SARS-CoV-2 (Sit et al. 2020) 1. Dogs have a low susceptibility to SARS-CoV-2 (Shi et al. 2020) Table 4. Origin of SARS-CoV-2.
-
According to the phylogenetic analysis of recently published SARS-CoV-2 genome data, SARS-CoV-2 is most closely related to the two SARS-like coronavirus sequences isolated from bats from 2015 to 2017, indicating that bat coronavirus and human SARS-CoV-2 share a common ancestor (Chan et al. 2020; Malik et al. 2020; Zhang LS et al. 2020). Bat coronavirus (RaTG13) has a short RdRp region whose genome sequence has 96.2% homology with SARS-CoV-2, providing evidence for the bat origin of SARS-CoV-2 (Zhou P et al. 2020). Additionally, SARS-CoV-2 is closely related to bat coronaviruses and has 100% amino acid similarity to BAT-SL-CoVZC45 in nsp7 and E proteins, suggesting that bats may be the reservoir host of SARS-CoV-2 or closely related viruses (Wu F et al. 2020). Additionally, Bayesian phylogeographic reconstruction shows that SARS-CoV-2 most likely originated from the bat SARS-like coronavirus circulating in the Rhinolophus bat family (Benvenuto et al. 2020). Recently, Jie Zhou et al. established and identified expandable intestinal organs derived from horseshoe bats of the Rhinolophus sinicus species that can recapitulate bat intestinal epithelium and found that the samples were fully susceptible to SARS-CoV-2 infection and sustained robust viral replication (Zhou J et al. 2020).
From SARS-CoV and MERS-CoV to SARS-CoV-2, all point to bats as the natural host of coronaviruses; however, many scholars are convinced that intermediate hosts facilitate the emergence of SARS-CoV-2 in humans (Chen Yun et al. 2020; Lu et al. 2020; Wan et al. 2020; Wrapp et al. 2020; Zhang LS et al. 2020). Roujian Lu et al. discussed several lines of evidence suggesting that other animals act as intermediate hosts between bats and humans (Lu et al. 2020). Firstly, the outbreak was first reported in late December 2019, when most bat species in the Wuhan hibernate. Secondly, no bats were sold or found at the Huanan seafood market, whereas various non-aquatic animals (including mammals) were available for purchase. Thirdly, the sequence identity between SARS-CoV-2 and its close relative bat-SL-CoVZC45 and bat-SL-CoVZXC21 is less than 90%, reflected in the relatively long branch between them. Hence, bat-SL-CoVZC45 and bat-SL-CoVZXC21 are most-likely not direct ancestors of SARS-CoV-2. Fourthly, in both SARS-CoV and MERS-CoV, bats act as primordial natural reservoir, while other animals (masked palm civet for SARS-CoV and dromedary camels for MERS-CoV) act as intermediate hosts. Below, we discuss intermediate, experimental, and agricultural hosts of SARS-CoV-2.
-
Many scientific research teams have compared the genomes of SARS-CoV-2 and Pangolin-CoV, finding the pangolin coronavirus genomes have more than 85.5% similarity to SARS-CoV-2 (Lam et al. 2020; Xiao et al. 2020; Zhang T et al. 2020). Kangpeng Xiao et al. published a very detailed study indicating that the amino acid identities of a coronavirus isolated from Malayan pangolins and SARS-CoV-2 concerning the E, M, N and S genes were 100%, 98.2%, 96.7%, and 90.4%, respectively. In particular, the receptor-binding domain of the pangolin-CoV S protein is nearly identical to that of SARS-CoV-2, except for one amino acid (Xiao et al. 2020). The discovery of multiple lineages of pangolin coronavirus and their similarity to SARS-CoV-2 suggest that pangolins may be the (or at least one) intermediate host of SARS-CoV-2 and should not be traded at wet markets to prevent zoonotic transmission.
However, another study came to opposite conclusions. Ping Liu et al. assembled the genomes of coronaviruses identified in sick pangolins, and their phylogenetic analyses do not support that SARS-CoV-2 arose directly from pangolin-CoVs (Liu P et al. 2020).
-
In intracellular parasites, the codon usage patterns resemble that of the host. Studies of the relative synonymous codon usage (RSCU) bias between viruses and their hosts suggest that viruses tend to evolve codon usage biases similar to that of their hosts (Bahir et al. 2009; Wang et al. 2016; Teesalu et al. 2009). The square of the Euclidean distance indicated that, SARS-CoV-2 has the highest similarity with snakes in terms of the RSCU bias as compared to bats, birds, marmots, humans, pangolins and hedgehogs. This may suggest that SARS-CoV-2 may use (or may have used) the snake translation mechanism more effectively than other animals (Zhou P et al. 2020).
However, Chengxin Zhang et al. disagreed with this conclusion because the study possessed several limitations (Zhang C et al. 2020). Junwen Luan et al. suggested that snakes should be ruled out from the potential host list for SARS-CoV-2 because the ACE2 of snake cannot associate with the viral S protein (Luan et al. 2020).
-
In their study (Liu Z et al. 2020), Zhixin Liu et al. used systematic comparisons and analyses to predict the interaction between the RBD of the S protein and its host receptor ACE2. Based on the interactions between key amino acids of the RBD and ACE2 and in agreement with above mentioned discussions pangolins and turtles may be a potential intermediate host for transmitting SARS-CoV-2 to humans. Additionally, considering Asn501 in the RBD domain and sites 41 and 353 of the ACE2 receptor, turtles and pangolins may be closer to humans than bats, indicating turtles and pangolins may have had the potential to serves as intermediate hosts of SARS-CoV-2.
However, Junwen Luan et al. found that the ACE2 of turtles cannot associate with S protein; thus, they should be ruled out from the potential host list for SARS-CoV-2 (Luan et al. 2020).
-
Some scholars used a tool called virus host prediction (VHP), developed based on a deep learning algorithm, to shed light on the host of SARS-CoV-2. They scored all the available vertebrate viruses in GenBank and filtered viruses with a similar infectivity pattern to SARS-CoV-2, whose hosts include canines, porcines, minks, tortoises, and felines. By comparing the infectivity patterns of all viruses infecting these vertebrates, they found that mink viruses show a closer infectivity pattern to SARS-CoV-2 and inferred that minks might be another candidate source of SARS-CoV-2 (Guo et al. 2020). Intriguingly, minks turned out to be highly permissive for SARS-CoV-2 (Enserink 2020). There was reported that minks is infected by SARS-CoV-2, with SARS-CoV-2 RNA in organ and swab samples, and one worker was assumed to have attracted the virus from mink (Oreshkova et al. 2020).
-
Ferrets are usually used as animal models of human respiratory tract virus infection. Jianzhong Shi et al. inoculated ferrets intranasally and analyzed SARS-CoV-2 in nasal irrigation and viral RNA in the rectal swabs of ferrets. The ferrets were euthanized on day 4 postinoculation, and their nasal turbinate, soft palate, tonsils, tracheas, lungs, heart, liver, spleen, kidneys, pancreas, small intestine, and brain were collected for viral RNA quantification and virus titration. SARS-CoV-2 was detected in the nasal turbinate, soft palate, and tonsils of all ferrets but not in any other organs tested, indicating SARS-CoV-2 can replicate in the upper respiratory tract of ferrets (Shi et al. 2020).
-
A serological study conducted among the cats in Wuhan observed the presence of SARS-CoV-2-neutralizing antibodies, indicating that cats can acquire SARS-CoV-2 infection under natural conditions (Zhang Q et al. 2020). Additionally, Jianzhong Shi et al. found that cats are highly susceptible to SARS-CoV-2 because antibodies against SARS-CoV-2 and the virus were detected in the viral RNA-positive nasal turbinate, soft palates, tonsils, tracheas and lungs of these cats (Shi et al. 2020). Furthermore, Peter J Halfmann et al. reported that infected cats can spread SARS-CoV-2 from one cat to another (Halfmann et al. 2020). Three domestic cats were inoculated with SARS-CoV-2 on day 0 and cohoused with three cats with no previous SARS-CoV-2 infection to assess on day 1. The virus was detected in all three cats cohoused with the inoculated cats. Therefore, a better understanding of the possible role of cats in the spread of SARS-CoV-2 to humans is needed to prevent COVID-19 pandemics. Interestingly, there is a report that tigers and lions, both belonging to the Felidae, tested positive for COVID-19 (https://abc7ny.com/bronx-zoo-tiger-with-coronavirus-tigers-lions/6122810/).
-
Two of 15 dogs in Hong Kong were infected with SARS-CoV-2 as indicated by virus RNA positivity, and the virus was isolated from the nose and mouth swabs of one animal (Sit et al. 2020). No evidence in that study showed that dogs could transmit infection to other dogs or people. Dogs have a low susceptibility to SARS-CoV-2 because viral RNA was detected in the rectal swabs of only two virus-inoculated dogs; however, viral RNA was not detected in any organs or tissues, and the dogs were all seronegative for SARS-CoV-2 (Shi et al. 2020).
Bats
Pangolins
Snakes
Turtles
Minks
Ferrets
Cats
Dogs
-
We have described the genome, structure, receptor, and origin of SARS-CoV-2. SARS-CoV-2 is the most pressing global health issue due to the ongoing pandemic. The identification of the natural host, the development of safe and protective vaccines as well as therapeutic drugs against SARS-CoV-2 are urgent measures to prevent the viruses from infecting more people in the future.
In general terms, the SARS-CoV-2-related disease outbreak once again proved the existence of virus reservoirs present in wild and domesticated animals, arguing for continuous surveillance and early warning programs.
-
This work is supported by the Fundamental Research Funds for the Central Universities (2020kfyXGYJ028), the National Natural Science Foundation of China (81861138044 and 91742114), the National Science and Technology Major Project (2017ZX10202203), and the Medical Faculty of the University of Duisburg-Essen and Stiftung Universitaetsmedizin, University Hospital Essen, Germany.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.














 DownLoad:
DownLoad: