HTML
-
Mumps is a common respiratory infection in children caused by mumps virus (MuV) (Muhlemann 2004). The primary symptoms of MuV infection are fever and swelling of the parotid gland and can also cause complications including orchitis, deafness, aseptic meningitis and encephalitis (Galazka et al. 1999). Since 1967, live attenuated vaccines have been developed for immunization against mumps in children, which significantly reduced the health, social and economic problems caused by mumps (Buynak and Hilleman 1966). Currently, live attenuated mumps vaccines widely used in the world include Jeryl-Lynn, S79, Urabe Am9, RIT 4385, Leningrad-3, Leningrad-Zagreb, Rubini, etc. (Rubin et al. 2015). Live attenuated S79 MuV vaccine which is originated from Jeryl Lynn strain has been licensed for vaccination since 1996 in China (Wu et al. 2018). Although live attenuated MuV vaccine is efficacious, it can rarely cause some undesirable adverse effects (Di Pietrantonj et al. 2020). In fact, many vaccinated children may experience side effects including fever, rash, lymphadenopathy, parotitis. In addition, the incidence of mumps is still high after a dose of measles-mumps-rubella (MMR) combination vaccine introduced in China (Cui et al. 2018; Ma C et al. 2018; Wu et al. 2018). Even vaccination coverage of the later second dose is high, it does not effectively prevent outbreaks of mumps among adolescents worldwide (Beleni and Borgmann 2018; Lewnard and Grad 2018; Ma R et al. 2018; Fields et al. 2019; Westphal et al. 2019). To solve the recurrence of mumps outbreaks, a proposal for a third dose of MMR vaccine against young adults has been proposed (Albertson et al. 2016; Marin et al. 2018). However, the third dose of MuV vaccine can control but not prevent outbreaks (Cardemil et al. 2017). Therefore, there is still an urgent need to develop more effective and safer MuV vaccines (May et al. 2018).
MuV is an enveloped, non-segmented negative-sense (NNS) RNA virus, belonging to the genus Orthorubulavirus, subfamily Rubulavirinae in the family Paramyxoviridae. The MuV genome encoded 7 transcription units: the nucleo (N), V/phospho/I (V/P/I), matrix (M), fusion (F), small hydrophobic (SH), hemagglutinin-neuraminidase (HN) and large (L) protein genes (Elango et al. 1988; Elliott et al. 1990).
The traditional method for developing live attenuated mumps vaccine is through continuous low-temperature passages in chicken embryo cells to identify an optimized viral strain with reduced virulence but sufficient immunogenicity (Buynak and Hilleman 1966). In addition, Japanese researchers developed several temperature-stable mutants among clinically isolated viral strains by nitrosoguanidine and ultraviolet light (Saika et al. 2006). However, these strategies are complicated and time-consuming and the resultant mutants are often unstable. The reverse genetics system provides a rapid and efficient strategy for rational design of live attenuated vaccines (Bukreyev et al. 1996; Xu et al. 2014; Nogales and Martinez-Sobrido 2016; Yang et al. 2016). The first MuV reverse genetics system was reported in 2000 (Clarke et al. 2000). As with other NNS RNA viruses (Garcin et al. 1995; Lawson et al. 1995; Jin et al. 1998), co-transfection of plasmids expressing viral anti-genome and support plasmids encoding viral nucleocapsid complex (N, P and L protein) was essential for the recovery of MuV. Using reverse genetics, researchers have shown that deletion of SH gene from MuV genome can further weaken the MuV vaccine strains and increase their immunogenicity. However, follow-up studies showed that this method might not be applicable to all vaccine strains (Xu et al. 2011). Therefore, exploration of new strategy to further improve the safety and efficacy is still needed.
The large (L) protein of NNS RNA viruses is the catalytic subunit of RNA dependent RNA polymerase that carries out replication and transcription (Whelan et al. 2004). During transcription, NNS RNA viruses produce mRNAs that are capped and methylated at 5′-end and polyadenylated at 3′-end (Poch et al. 1990). The mRNA capping and methylation are catalyzed by the L protein. Specifically, addition of mRNA cap is accomplished by the conserved V (CR V) of L protein. The mRNA cap structure is further methylated by guanine-N-7 (G-N-7) and ribose 2′-O methyltransferases (MTases) which are located in the CR Ⅳ of L protein (Li et al. 2006). Recently, cryo-EM structure of L proteins of two NNS RNA viruses, vesicular stomatitis virus (VSV) and human respiratory syncytial virus (RSV), has been solved (Ge et al. 2010; Gilman et al. 2019).
mRNA cap G-N-7 methylation is essential for viral protein translation (Muthukrishnan et al. 1976) whereas ribose 2′-O methylation is associated with viral evasion of host interferon (IFN)-mediated innate immune responses (Zust et al. 2011). Therefore, mRNA cap methylation is an attractive target for development of antiviral drugs and live attenuated vaccines. Because virus lacking mRNA cap G-N-7 methylation will reduce viral protein translation, which in turn down-regulate replication and transcription, leading to virus attenuation. Viruses lacking mRNA cap ribose 2′-O methylation will make the virus more easily recognized by host cells, activate the interferon system and then promote T cell immune response and B cell immune response and improve immunogenicity. This finding has been proven in several NNS RNA viruses, including measles virus (MV), human metapneumovirus (hMPV), avian metapneumovirus (aMPV), rabies virus (RABV) and vesicular stomatitis virus (VSV) (Ma et al. 2014; Sun et al. 2014; Zhang et al. 2014; Wang et al. 2018). These studies showed that recombinant viruses lacking MTase activity were highly attenuated both in vitro and in vivo yet retained high immunogenicity.
We hypothesized that reducing MTase activity can further improve the safety and immunogenicity of mumps vaccines. Using the robust reverse genetics system of Chinese S79 vaccine strain (MuV-S79) established in our previous research (Zhou et al. 2019b), we generated a total of eight recombinant MuVs with amino acid substitutions in the MTase domain of the L protein. These rMuVs mutant strains were significantly more attenuated in cell culture compared with the parental vaccine strain and were genetically stable when passing repeatedly in cell culture for 10 passages. In addition, we found that three rMuV mutants were significantly more attenuated in cell culture and in cotton rats yet retained wild-type level of immunogenicity. We also identified one rMuV mutant that was replicated as efficiently as the parental vaccine strain but had a higher immunogenicity. Collectively, our results suggest that rMuV carrying mutations in the SAM binding site or MTase catalytic site may have improved safety and/or immunogenicity.
-
BHK-SR19-T7 cells (kindly offered by Apath, LLC, Brooklyn, NY), Vero cells (African green monkey kidney, ATCC-CCL-81) and Hela cells (kindly offered by Wangxing Hu, Ph.D., The Second Affiliated Hospital of Zhejiang University School of Medicine) were grown in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 10% fetal bovine serum (FBS, Life Technologies, USA). To screen BHK-SR19-T7 cells expressing T7 polymerase, the medium of the cells was supplemented with 10 μg/mL puromycin (Life Technologies) during passages.
The MuV-S79 vaccine strain (kindly provided by Professor Yiyu Lu, Zhejiang CDC, China) and MuV wild-type strain (isolated from clinical samples in Children's Hospital, Zhejiang University School of Medicine, genotype F) were passaged in Vero cells. Recombinant rMuV-S79-A1814G, rMuV-S79-G1816A, rMuV-S79-G1818A, rMuV-S79-D1892A, were mutated from the methyltransferase binding site of rMuV-S79. And rMuV-S79-K1792A, rMuV-S79-D1917A, rMuV-S79-K1953A, rMuV-S79-E1990A, were mutated from the methyltransferase catalytic site of rMuV-S79. Viruses were handled in BSL-2 laboratory adhered to standard biosecurity and institutional safety procedures.
-
23 clinical samples of mumps-positive throat swabs were used to isolate and identify the wild type virus. RT-PCR tests were performed on clinical mumps throat swab samples. 100 μL of positive clinical mumps samples were inoculated into Vero cell monolayers and adsorbed at 37 ℃ for 1 h. The Vero cells were discarded the adsorbed liquid and added 2 mL of DMEM medium containing 2% FBS and cultured at 37 ℃. Cytopathic effects (CPE) were observed and recorded daily. If no CPE was observed, the supernatant was harvested 7 days after incubation and blindly passaged three times on a new Vero cell monolayer. When significant CPE was observed, the cells were frozen and thawed three times to harvest the viral supernatant and stored at - 80 ℃. The isolated virus was plaque purified and confirmed by RT-PCR and used as the MuV wild-type strain in the study.
-
rMuVs mutants were obtained by specific amino acid mutations in the CR Ⅳ of rMuV-S79 L protein. Using the PFU Site-Directed Mutagenesis Kit (NEB) and using rMuV-S79 (GenBank: MT732483) as a template, the key amino acids (K1792, G1816, G1818, D1892, D1917, K1953, E1990) were mutated to alanine by primers (Supplementary Table S1). And A1814 was mutated from alanine to glycine. The mutation was confirmed by sequencing. These plasmids were named pYES-MuV-S79-K1792A, pYES-MuV-S79-A1814G, pYES-MuV-S79-G1816A, pYES-MuV-S79-G1818A, pYES-MuV-S79-D1892A, pYES-MuV-S79-D1917A, pYES-MuV-S79-K1953A and pYES-MuV-S79-E1990A.
-
The full-length MuV clone pYES-MuV-S79 (12.5 μg) and its supporting plasmids pT7-MuV-S79-NP (3.75 μg), pT7-MuV-S79-P (3.75 μg), pT7-MuV-S79-L (1.25 μg) were co-transfected into 70%-80% confluent BHK-SR-19-T7 cells in T25 cell culture flask. The transfection was operated by using Lipofectamine 2000 and referring to the manufacturer's instructions. After 6 h of incubation at 37 ℃, fresh Opti-MEM culture was added to replace the old medium. At 72 h post-transfection, transfected cells were scraped off, resuspended and co-cultured with 75% confluent Vero cells in T25 cell culture flask at 37 ℃ with 5% CO2 for 3-7 days until mumps-induced specific cytopathic effect was observed. Each recombinant virus was plaque isolated.
-
24-well Vero cells (5 × 104 /well) were grown overnight for being infected with rMuVs. After 1 h of adsorption, the virus adsorption solution was discarded and the cells were washed twice with PBS and added with 0.5 mL DMEM containing 2% FBS. After 48 h, cells were washed twice with PBS and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature (RT), then ruptured with 0.2% Triton X-100 in PBS for 20 min and blocked with 2% BSA in PBS for 1 h. Further, the treated cells and the specific mouse anti-mumps N protein specific antibody (ab9876, Abcam, 1:20) were incubated for 1 h at room temperature, following incubation with anti-mouse IgG (A21203, Alexa Fluor ® 594 donkey, Invitrogen 1:2000) for 0.5 h at RT. Finally, the nucleus was stained by DAPI (1:10, 000) and incubated for 5 min in the dark; and the results were observed under a fluorescence microscope.
-
After discarding the medium and washing the cells with PBS, Vero cells in 6-well plates were inoculated with rMuVs at a multiplicity of infection (MOI) of 0.1. The virus was collected per 24 h by freezing and thawing the cells three times and centrifuged at 4000 g, 4 ℃ for 25 min. The cell lysate was denatured at 100 ℃ for 5 min and analyzed on a 10% polyacrylamide bis-Tris gel. The separated protein was transferred to a polyvinylidene difluoride membrane. The membranes were blocked with 5% skim milk in PBS and subsequently incubated with specific mouse anti-mumps N protein specific antibody (ab9876, Abcam, 1:100) for 1 h at RT, following incubation with anti-mouse IgG (7076S, CST, 1:3000) for 0.5 h at RT. Finally, the membranes and ECL were incubated (20-50-120, Biological Industries) for 2 min and then developed.
-
Prepare 100 mL of the covering medium one day in advance, components: 3.2% aqueous solution of sodium carboxymethylcellulose 25 mL (autoclave for 30 min), 75 mL DMEM medium containing 2% FBS and 1% PS. Discard the medium of Vero cells in six-well plates at a density of 5 × 105 cells per well and wash the cells with PBS before adsorption of serial dilutions of mumps. After shaking in constant temperature for 1 h, virus adsorption solution was discarded and cells were covered with 2 mL of the covering medium per well. Then 6-well plates were placed at 37 ℃, 5% CO2 cell incubator for 7 days. After 7 days, cells were fixed 2 mL of 4% formaldehyde for 2 h per well. The plaques formed by the mumps virus infection were visualized by staining with 0.05% (wt/vol) crystal violet each well for 15 min. Calculate the virus titration by counting the number of plaques in the cell and combining the dilution gradient.
-
The medium of Vero cells was discarded and the cells were washed with PBS. Vero cells were adsorbed 100 PFU of each virus per well. After shaking in constant temperature for 1 h, virus adsorption solution was discarded and cells were covered with 2 mL of the covering medium per well. Then 6-well plates were placed in cell incubator at 37 ℃ with 5% CO2 for 7 days. After 7 days, cells were fixed with 2 mL of 4% formaldehyde for 2 h per well. The plaques formed by the mumps virus infection were visualized by staining with 0.05% (wt/vol) crystal violet each well for 15 min. The bottoms of the six-well plate were scanned after crystal violet staining and the diameter of the virus plaque was measure by the 'ruler' tool in the PS software. 10-15 plaques were randomly picked to measure their diameter and averaged as the final size of the plaque.
-
After discarding the medium and washing the cells with PBS, Vero/Hela cells in 6-well plates were inoculated with rMuVs at a multiplicity of infection (MOI) of 0.1. The virus was collected per 24 h by freezing and thawing the cells three times and centrifuged at 4000 g, 4 ℃ for 25 min. Virus titers were determined by plaque assays as described above (Virus titration). The experiment was repeated for 3 times to plot growth curves.
-
Inoculation of Vero cells in T25 flasks with mRNA methyltransferase-deficient mumps virus vaccine strain or rMuV-S79 at MOI = 0.1. After virus infection of Vero cells for 1 h, the virus adsorption solution was discarded, then the cells were washed with PBS and 4 mL of DMEM medium containing 2% FBS was added to each well and cells were placed in a 37 ℃ cell culture incubator. After CPE reaches 80% of the cell monolayer, the virus was collected by freezing and thawing the cells three times and centrifuged at 4000 g, 4 ℃ for 25 min. The collected virus was used for the next passage. Repeat the passage 10 times as described above. Inoculation of primary chicken embryo fibroblast cells (CEF) in T25 flasks with mRNA methyltransferase-deficient mumps virus vaccine strain or rMuV-S79 at MOI = 1. After 72 h, the virus was collected by freezing and thawing the cells three times and centrifuged at 4000 g, 4 ℃ for 25 min. The collected virus was used for the next passage. Repeat the passage 3 times as described above.
The entire genome of each recombinant virus was amplified by RT-PCR and sequenced. The genomic information of each methyltransferase-deficient rMuVs was amplified by RT-PCR and sequenced.
-
Viral RNA was extracted by using RNeasy mini-kit (Qiagen) according to the manufacturer's instructions. A 950 bp DNA fragment of CR Ⅳ derived from MuV L protein was amplified by a One-Step RT-PCR kit (Qiagen) according to the manufacturer's instructions using primers MuV-13722F and MuV-14671R (Supplementary Table S1). To confirm that the designed mutation was indeed present in the CR Ⅳ of the MuV L protein, the PCR product was sequenced using the primers described above.
-
Ten T150 flasks of more than 90% confluent Vero cells were adsorbed with mRNA methyltransferase-deficient mumps virus with MOI = 0.1. After 80% of cytopathic effects (CPE) were observed, the virus was collected by freezing and thawing the cells three times and centrifuged at 4000 g, 4 ℃ for 25 min. Then the virus was pelleted by ultracentrifugation at 25, 000 g for 2 h and resuspend in 500 μL of DMEM containing 10% Trehalose. The ultracentrifuge tube was placed on ice overnight to completely dissolve the suspended virus. Purified virus was aliquoted and stored at -80 ℃ for subsequent animal testing the next day.
-
Female specific-pathogen-free (SPF) cotton rats (kindly provided by Professor Enmei Liu from Children's hospital of Chongqing Medical University), 4-5 weeks old, were used in the experiment. The experimental animals were housed within the facilities of the Children's Hospital of Chongqing Medical University. During the experiment, cotton rats were anesthetized using isoflurane and sacrificed by carbon dioxide asphyxiation.
-
In this animal experiment, the virulence assay of mRNA methyltransferase-deficient mumps virus vaccine strain was performed. Forty-five 4 weeks old, female SPF cotton rats were randomly divided into 9 groups: rMuV-S79, rMuV-S79-K1792A, rMuV-S79-G1816A, rMuV-S79-G1818A, rMuV-S79-D1892A, rMuV-S79-D1917A, rMuV-S79-K19538A, rMuV-S79-E1990A and DMEM. Isoflurane anesthetized cotton rats were inoculated intranasally at a dose of 1.0 × 106 PFU. After inoculation, clinical signs, weight and mortality were observed and recorded accordingly. On the 4th day after inoculation, cotton rats were euthanized and lung, spleen and brain tissue samples were collected. The collected left lungs, left brain and spleen tissues were weighed and then added to 1 mL of pre-cooled DMEM for grinding and homogenization, then centrifuged at 4000 g for 4 min at 4 ℃. The supernatant was collected and stored in a new pre-cooled sterile tube for plaque assay. For histopathological detection, the right lung and right brain tissues were fixed in 4% formaldehyde. Fixed tissues were embedded in paraffin and sectioned at 5 μm. Slides were then stained with hematoxylin-eosin (H&E) for the examination of histological changes by light microscopy.
-
In this animal experiment, the immunity assay of constructed vaccine strains was performed on forty-five 4-week-old female SPF cotton rats by same grouping method as above. Isoflurane anesthetized cotton rats were inoculated intranasally at a dose of 1.0 × 106 PFU rMuVs and the serum of the cotton rat was collected at the 2nd, 3rd, 4th, 5th, 7th and 9th week after inoculation for antibody detection. The animals were challenged with wild-type MuV strain by intranasal inoculation with a dose of 1.0 × 107 PFU/100 μL and then the morbidity and mortality of cotton rats were observed daily. Four days after challenge, all cotton rats were euthanized and lung tissues were collected for detection of intrapulmonary virus titers and histopathological research as described above.
-
First, the tissue samples were fixed with 4% formaldehyde and embedded in paraffin. The slices were cut into 4 μm thick slices and then incubated at 60℃ for 2 h. Subsequently, the slides were de-waxed and then put into a beaker filled with repair liquid and then put in a pressure cooker to boil. After heating at the set pressure for 2 min, the slides were cooled by water. Then The sections were incubated in 3% H2O2 solution about 10 min to block endogenous peroxidase activity. The slides were incubated with normal goat serum (1/20 dilution in PBS) at 37 ℃ for 15 min and subsequently with mouse anti-mumps N protein mAb (Abcam, ab9880, 1/500 dilution in PBS) at 4 ℃ overnight. Afterward, the slides were cultivated with a biotinylated goat anti-mouse HRP-IgG (ZSGB-BIO, PV-8000, 1/1) at 37 ℃ for 30 min. The slides were then developed using a diaminobenzidine (DAB) for 2 min and hematoxylin for 20 s as a counterstain.
-
Mumps virus-specific neutralizing antibody titers in cotton rat serum were detected by plaque reduction neutralization assay. The collected serum samples were inactivated in a constant temperature water bath at 56 ℃ for 30 min. Then the inactivated serum samples were serially diluted with DMEM medium in 96-well plates (1:16 to 1:2048) mixed with an equal volume of DMEM containing approximately 100 PFU/well rMuV-S79 and incubated for 1 h at 37 ℃ constant temperature shaker. The virus-serum mixture was adsorbed to single-layer Vero cells in 6-well plates. Further virus adsorption solution was discarded and cells were covered with 2 mL of the covering medium (as virus titration) per well, then 6-well plates were placed in cell incubator at 37 ℃ with 5% CO2 for 7 days. After 7 days, cells were fixed with 2 mL of 4% formaldehyde per well for 2 h. The plaques formed by the mumps virus infection were visualized by staining with 0.05% (wt/vol) crystal violet for 15 min. The plaques were counted and 50% plaque reduction titers were calculated as the MuV-specific neutralizing antibody titers.
-
Sequence data of rMuVs that support the findings of this study have been deposited in GenBank with the accession numbers: rMuV-S79, MT732483; rMuV-S79-K1792A, MT732484; rMuV-S79-A1814G, MT732485; rMuV-S79-G1816A, MT732486; rMuV-S79-G1818A, MT732487; rMuV-S79-D1892A, MT732488; rMuV-S79-D1917A, MT732489; MuV-S79-K1953A, MT732490; rMuV-S79-E1990A, MT732491. The wild type mumps virus GenBank accession number: KF170916.
-
Statistical analysis of data was performed using two-way ANOVA of Prism, version 8.0. P values (P) Less than 0.05 were considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Cells and Viruses
Specimen and Virus Separation
Site-directed Mutagenesis
Recovery of Infectious rMuV-S79 from Transfected Cells
Immunofluorescence Assay
Western Blot
Virus Titration
Virus Plaques Size
Recombinant Virus Replication Kinetics in vitro
Genetic Stability of rMuV-S79 Mutants in vitro
Confirmation of Sequences by RT-PCR
Purification of rMuV-S79
Animal Experiments
Replication of rMuV-S79s in Cotton Rats
Immunogenicity of rMuV-S79s in Cotton Rats
Immunohistochemical (IHC) Staining
Detection of Mumps Virus Neutralizing Antibody Levels
Sequence Data of rMuVs and Wild Type MuV
Statistical Analysis
-
We first performed a sequence alignment of the CR Ⅳ (MTase region) of the L proteins among NNS RNA viruses (Fig. 1). An MTase catalytic site (K-D-K-E tetrad) is conserved in the CR Ⅳ of all L proteins. In the methylation reaction, the MTase must bind to the methyl donor, SAM, in order to achieve methylation. A SAM binding site (GxGxG…D motif) is conserved in the L proteins of NNS RNA viruses. However, the SAM binding site of the L protein of MuV, Human parainfluenza virus-2 (HPIV2) and Newcastle disease virus (NDV) contains a naturally occurring mutation. Specifically, the first amino acid residue in the SAM biding site is an alanine but not a glycine. Sequence alignment indicated that amino acids including A1814, G1816, G1818 and D1892 responsible for SAM binding in MuV L protein; and amino acids K1792, D1917, K1953 and E1990 correspond to the catalytic K-D-K-E tetrads of the MuV L protein. We performed an alanine scanning mutagenesis in the SAM binding site and K-D-K-E motif in MuV L protein and each of these amino acid residues was mutated to alanine. Since the first amino acid of the SAM binding site is naturally an alanine (A1814), we mutated this alanine to glycine to create a SAM binding site which is conserved in the L proteins of most NNS RNA viruses. Primers used for mutagenesis are listed in Supplementary Table S1. Each of these mutations was introduced into plasmid pYES-MuV-S79 encoding a full-length cDNA clone of MuV vaccine strain S79.
Figure 1. Sequence alignment in the conserved region VI (CR Ⅳ) of the L proteins of NNS RNA viruses. The sequence of CR Ⅳ in the L protein was selected and compared using the Clustal Omega software. The coordinates were based on the amino acids 1790-1990 of the L protein of MuV-S79. The predicted SAM binding sites (GxGxG…D) are shown in orange and methyltransferase catalytic sites (K-D-K-E) are shown in grey. Representative members in the Paramyxoviridae (MuV, mumps virus; MeV, Measles virus; HeV, Hendra virus; NDV, Newcastle disease virus; CDV, canine distemper virus; PPRV, peste des petits ruminants virus; SeV, Sendai virus; RPV, Rinderpest virus; HPIV, human parainfluenza virus-2), Pneumoviridae (PVM, pneumonia virus of mice; HMPV, human metapneumovirus; BRSV, bovine respiratory syncytial virus; HRSV, human respiratory syncytial virus), Rhabdoviridae (VSV, vesicular stomatitis virus) were selected for sequence alignment.
-
Using the reverse genetics system, all eight rMuVs were recovered, which were named rMuV-S79-K1792A, rMuV-S79-A1814G, rMuV-S79-G1816A, rMuV-S79-G1818A, rMuV-S79-D1892A, rMuV-S79-D1917A, rMuV-S79-K1953A and rMuV-S79-E1990A. MuV-induced CPE were observed after 2-4 days (Fig. 2A). Subsequently, MuV NP protein was detected in Vero cells infected by each recombinant virus using an immunofluorescence assay (IFA) and Western blot (Fig. 2B and Supplementary Fig. S1A). The parent strain obtained the top NP antigen protein expression at 72 h after transfection, while rMuVs were delayed by 1-3 days (Supplementary Fig. S1B). The top NP antigen protein expression of rMuV-S79-D1917A and rMuV-S79-E1990A are similar to the parent virus, while the top expression of other rMuVs is lower than the parent virus (Supplementary Fig. S1C). The sizes of virus-induced plaques differed between the rescued parental and mutant viruses (Supplementary Fig. S2). Each recombinant virus was plaque purified, as demonstrated in Fig. 3 and 4A, rMuV-S79-K1792A (0.64 ± 0.11 mm), rMuV-S79-G1816A (0.66 ± 0.10 mm), rMuV-S79-G1818A (0.52 ± 0.13 mm), rMuV-S79-D1892A (0.18 ± 0.09 mm) and rMuV-S79-D1917A (0.48 ± 0.10 mm) formed significantly smaller plaques in diameter compared to the parental rMuV-S79 (0.82 ± 0.16 mm). Interestingly, rMuV-S79-A1814G (1.23 ± 0.18 mm) formed bigger plaques than the parental virus. Recombinant rMuV-S79-K1953A (0.85 ± 0.12 mm) and rMuV-S79-E1990A (0.70 ± 0.13 mm) had similar plaque sizes with the parental rMuV-S79 strain. Finally, the genome of each recombinant virus was amplified by RT-PCR and sequenced (Supplementary Fig. S3). The results showed that each rMuV contained only the designed mutation.
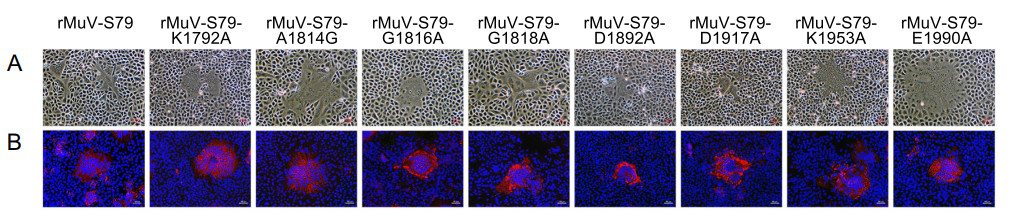
Figure 2. Rescue recombinant MuV mutants using reverse genetics. BHK-SR-19-T7 cells were co-transfected with a plasmid encoding full-length MuV antigenome and supporting plasmids encoding N, P and L genes. At day 3 post-transfection, cells were harvested and co-cultured with Vero cells for 24 to 96 h. Cell culture supernatants were used for next passage in Vero cells. A Specific cytopathic effects of rescued mumps virus on Vero cells. Scale bar: 50 μm. B Indirect immunofluorescence analysis of MuV rescued by Vero cells at 48 h post infection, red is labeled as mumps N protein and blue is labeled as Vero cell nucleus. Scale bar: 50 μm.
Figure 3. Plaque morphology of rMuVs. A plaque assay was performed in Vero cells for each recombinant virus. After 7 days of incubation, plaques were fixed and stained with crystal violet.
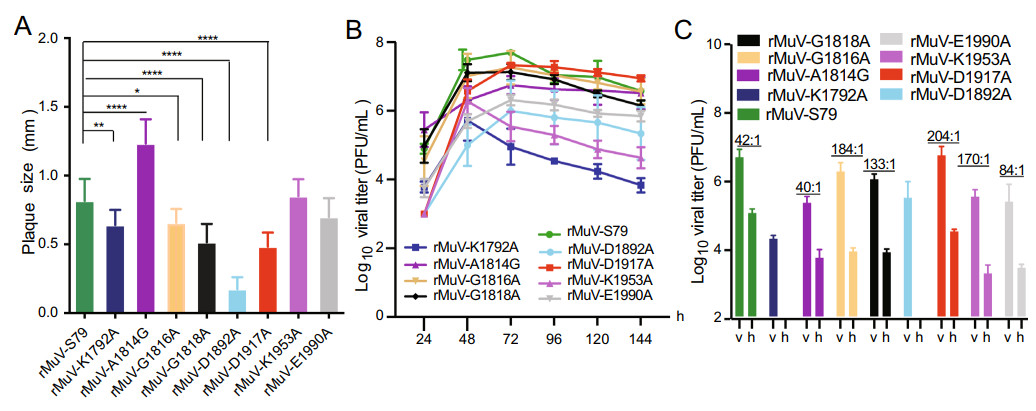
Figure 4. Growth characteristics of rMuV mutants. A Comparison of plaque size of recombinant virus. Fifteen plaques of each recombinant virus were measured and the average plaque diameter was calculated. *P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001. B Growth kinetics of rMuV mutants. Vero cells were infected by each recombinant virus at an MOI of 0.1, virus was harvested at the indicated time points. Virus titers of the rMuV mutants were determined by multiple step growth curve plaque assay in Vero cells. C Replication of rMuVs in IFN-/IFN + cell lines. Confluent Vero (v) and Hela cells (h) were infected with rMuVs at an MOI of 0.1. After 72 h post infection, virus titers were determined by plaque assay.
-
We evaluated the replication kinetics of these recombinant MuV mutants in Vero cells (Fig. 4B). The virus growth of most rMuV-S79 mutants was delayed by 1 day compared with the parental rMuV-S79. However, rMuV-S79-A1814G replicated earlier than the parental rMuV-S79 and reached a peak titer 6.75 ± 0.26 log10PFU/mL. The peak titers of rMuV-S79-K1792A and rMuV-S79-D1892A were 5.7 ± 0.59 log10PFU/mL and 6.0 ± 0.89 log10PFU/mL, respectively, which were lower than that of rMuV-S79 (7.6 ± 0.22 log10PFU/mL). The rMuV-S79-K1953A and rMuV-S79-E1990A, with a similar plaque size as rMuV-S79, had a lower peak titer (5.55 ± 0.43 log10PFU/mL and 6.32 ± 0.15 log10PFU/mL) than that of the parental strain. The peak titer of rMuV-S79-D1917A was 7.33 ± 0.06 log10PFU/mL, which was comparable to the parental strain at 72 h after infection.
Next, we determined whether defects in rMuVs replication are cell type specific. Briefly, Vero (IFN-) and Hela (IFN +) cells were infected with rMuVs and virus titers were determined in 72 h after infection by plaque assay. As shown in Fig. 4C, the ratio of viral titer between rMuV-S79 in Vero and Hela cells was 42:1. The ratio of viral titer between rMuV-S79-A1814G in Vero and Hela cells was 40:1 similar to the parental strain. The ratios of viral titer between rMuV-S79-G1816A, rMuV-S79-G1818A, rMuV-S79-D1917A, rMuV-S79-K1953A and rMuV-S79-E1990A in Vero and Hela cells were 184:1, 133:1, 204:1, 170:1 and 84:1 respectively, which higher than that of the parental strain. rMuV-S79-K1792A and rMuV-S79-D1892A cannot be detected > 100 PFU/mL viral titer in Hela cells. These data indicated that the replication kinetics of rMuVs in Hela cells was reduced and the reduction of rMuVs was more obvious than the parent strain.
The parental rMuV-S79 induced significant CPE in Vero cells at 72 h post-inoculation (hpi). All rMuV-S79 mutants except rMuV-S79-A1814G had significantly delayed CPE comparing with the rMuV-S79 (Fig. 5). Recombinant rMuV-S79-A1814G developed an earlier CPE, formed large syncytia at 24 hpi and reached a maximum CPE at 48 hpi. These data showed that rMuV-S79-A1814G had an increased syncytial formation whereas all other rMuV-S79 mutants had a significant attenuation in viral replication compared to the parental rMuV-S79.
-
We then tested the genetic stability of these recombinant viruses in cell culture. All rMuV-S79 mutants were passaged 10 times in Vero cells. At each passage, the MTase region in the L gene was amplified by RT-PCR and sequenced. The results showed that the L gene in each passage retained the desired mutation. At passage 10, the entire genome of each rMuV-S79 mutant was sequenced. All viruses retained the desired mutations and no other mutations were found in the viral genome as showed in Supplementary Fig. S3. The mumps vaccines are produced in primary chicken embryo fibroblast cell (CEF), so we tested the genetic stability of these rMuVs in CEF cells. rMuVs carrying mutations were repeatedly passaged 3 times in CEF cell and the entire genome of each recombinant virus was amplified by RT-PCR and sequenced. No mutations were found in the genome. These results indicated that these rMuV-S79 mutants were genetically stable in cell culture.
-
We next tested the replication ability of these rMuV mutants in vivo. Briefly, five-week-old, specific-pathogen-free (SPF) cotton rats were anesthetized with sevoflurane and inoculated intranasally with 1 × 106 PFU of each rMuV-S79 mutant (except rMuV-S79-A 1814G) or the parental rMuV-S79. No weight loss, death or obvious clinical signs of respiratory infection was observed in cotton rats. At day 4 post-inoculation, cotton rats were euthanized, lung, spleen and brain tissues were collected for virus titration and histology. No infectious MuV was detected in spleen and brain tissues for all groups. Viral titer in the lung tissues of rMuV-S79-G1818A-infected cotton rats was 2.97 ± 0.15 log10 PFU/g (P > 0.05), which is similar to rMuV-S79-infected control group (2.75 ± 0.12 log10 PFU/g). Viral titer in the rMuV-S79-G1816A (2.25 ± 0.06 log10 PFU/g, P < 0.001) and rMuV-S79-E1990A groups (2.48 ± 0.10log10 PFU/g, P < 0.05) were significantly lower than that in the rMuV-S79 group. However, no infectious virus was detected in the lungs of rMuV-S79-K1792A, rMuV-S79-D1892A and rMuV-S79-K1953A groups (Table 1). Also, no significant histologic lesion was detected in the lung and brain tissues for all groups (Supplementary Fig.S4A, S6A). To determine the viral antigen distribution in lung and brain tissues, IHC was performed using an antibody against the MuV N protein. As shown in Fig. 6, viral antigen-positive cells were detected inside the alveolar cells in lungs of rMuV-S79-infected cotton rats. Interestingly, the virus antigen-positive cells were detected in the bronchiolar cells of the rMuV-K1792A, rMuV-D1892A and rMuV-E1990A infected group, which had lower viral replication kinetics. Fewer antigen-positive cells were detected at the alveolar cells in lung tissues of other rMuVs infected groups. No viral antigen was detected in brain tissues from all groups as shown in Supplementary Fig. S5A. These results showed that rMuV mutants were more attenuated in replication in cotton rats compared to the rMuV-S79 virus strain.
Group Viral titer in lung Percentage of infection Viral titer (log10 PFU/g) rMuV-S79 80 2.75 ± 0.12 rMuV-S79-K1792A 0 ND rMuV-S79-G1816A 100 2.25 ± 0.06 rMuV-S79-G1818A 80 2.97 ± 0.15 rMuV-S79-D1892A 0 ND rMuV-S79-D1917A 80 3.04 ± 0.10 rMuV-S79-K1953A 0 ND rMuV-S79-E1990A 100 2.48 ± 0.10 DMEM 0 ND ND: not detected; Data each group were the mean of 5 cotton rats ± SD Table 1. Infectivity of rMuV mutants in cotton rats.
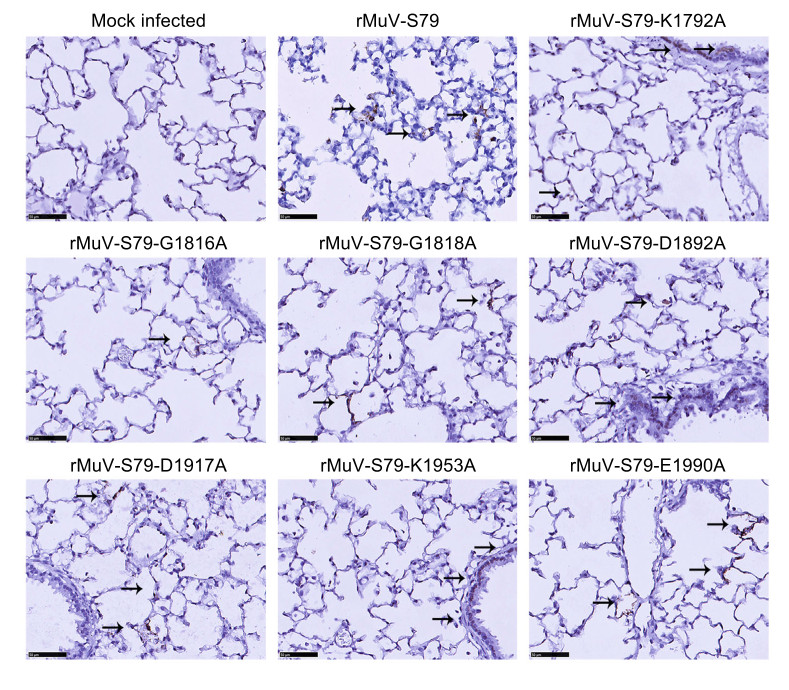
Figure 6. Immunohistochemical (IHC) staining of lungs of cotton rats infected with rMuV mutants. Cotton rats were intranasally inoculated with 1.0 × 106 PFU of each rMuV mutant and were sacrificed at day 4 post-inoculation. The right lung from each cotton rat (N = 45) was fixed with 4% formaldehyde and embedded in paraffin, sectioned at 4 μm and stained with monoclonal antibody against mumps N protein (Abcam, ab9880, 1/500 dilution in PBS) to determine the distribution of viral antigen. Arrows, antigen-positive cells. Scale bar: 50 μm.
-
Next, the immunogenicity of rMuV-S79 mutants was determined in cotton rats. Five-week-old SPF cotton rats were anesthetized with sevoflurane and inoculated intranasally with 1 × 106 PFU of each rMuV-S79 mutant or parental rMuV-S79 and serum samples were collected weekly for determination of neutralizing antibody titer. At week 4 post-vaccination, each group was challenged with 1.0 × 107 PFU of a wild type MuV strain. At 4 days post-challenge (dpc), cotton rats were euthanized; and lung tissues were collected for virus titration and histologic examination.
We compared the dynamics of neutralizing antibody responses after vaccination with each rMuV mutant. Figure 7A shows cotton rats vaccinated with the parental vaccine strain rMuV-S79 induced neutralizing antibodies at week 2 post-vaccination, reached a peak titer at week 3 and started to decline at week 4. Antibody titers in rMuV-S79-K1792A, rMuV-S79-D1892A, rMuV-S79-G1818A and rMuV-S79-K1953A were not detectable at week 2, suggesting a significant delay in antibody responses for those mutants. Recombinant rMuV-S79-K1792A and rMuV-S79-D1892A had significantly lower antibody titers than the rMuV-S79 group over the entire time period. Although rMuV-S79-G1818A and rMuV-S79-K1953A had a significant delay in antibody responses (P < 0.05) but reached a comparable peak titer with the rMuV-S79 group. Recombinant rMuV-S79-E1990A induced similar antibody responses to rMuV-S79 (P > 0.05). Importantly, neutralizing antibody titers in the rMuV-S79-D1917A group triggered significantly higher antibody titers in all time points comparing with the parental rMuV-S79 group (P < 0.05). Thus, these data demonstrated that rMuV-S79 mutants had variable impacts on antibody responses. Their ability of triggering neutralizing antibodies can be ranked as following: rMuV-S79-D1917A > rMuV-S79, rMuV-S79-E1990A > rMuV-S79-G1816A, rMuV-S79-G1818A > rMuV-S79-K1953A > rMuV-S79-K1792A, rMuV-S79-D1892A.
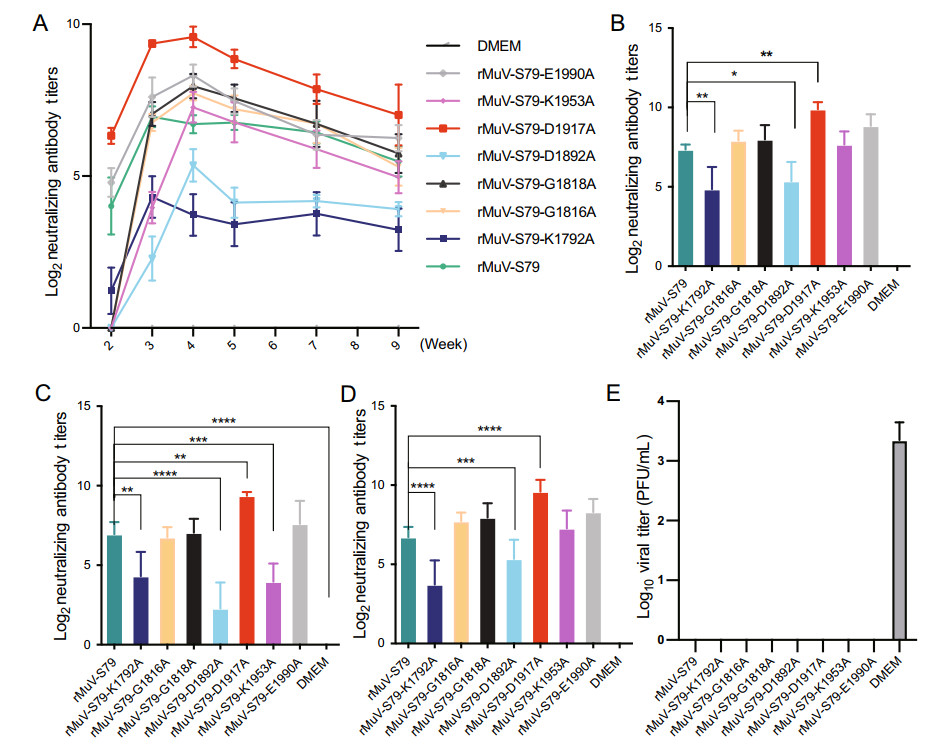
Figure 7. Neutralizing antibody titers triggered by rMuVs in cotton rats. A Dynamics of neutralizing antibody titers triggered by each recombinant MuV in cotton rats. The serum of the cotton rats was collected at 2, 3, 4, 5, 7 and 9 weeks after immunization and antibody titers were determined using a plaque reduction neutralization assay. Data each group were the mean of 5 cotton rats ± SEM. B Comparison of the highest neutralizing antibody titers induced by each rMuVs in cotton rats. C Comparison of neutralizing antibodies at week 3. D Comparison of neutralizing antibodies at week 4. E Protective ability of rMuV-S79 mutants in cotton rats. *P < 0.05; **P < 0.01; ***P < 0.001.
The peak antibody titer of the rMuV-S79-D1917 group was higher than that of the parental strain, however, the rMuV-S79-K1792A and rMuV-S79-D1892A groups had lower peak antibody titers than the parental strain; and the other groups were similar to the rMuV-S79 group (Fig. 7B). Importantly, neutralizing antibody titer in the rMuV-S79-D1917A group produced antibodies earlier and higher than that of the parental vaccine group (P < 0.05). We also analyzed the peak antibody titer either at week 3 or 4 post-immunization. At week 3, rMuV-S79-K1792A, rMuV-S79-D1892A and rMuV-S79-K1953A produced significantly lower neutralizing antibodies than the parental strain whereas rMuV-S79-D1917 produced a higher neutralizing antibody titer (Fig. 7C). At week 4, rMuV-S79-D1892A and rMuV-S79-K1953A produced antibodies similar to the rMuV-S79, while rMuV-S79-D1917 and rMuV-S79-K1792A had the same trend as week 3 (Fig. 7D).
For the unvaccinated challenged control group, an average virus titer of 3.35 ± 0.26 log10 PFU/g was detected in the lungs (Fig. 7E). In contrast, no MuV titer was detected in the lungs from cotton rats vaccinated with each rMuV mutant and challenged with wild type MuV. These results showed that all rMuV-S79 mutants provided complete protection against MuV replication in the lungs. No significant lung and brain histology lesion was found in all groups, even in the unvaccinated cotton rats (Supplementary Fig. S4B, S6B). A few of inflammatory cell infiltrations were found in the lung of cotton rats immunized with rMuVs.
For lung tissues from unvaccinated controls challenged with wild type MuV, viral antigens were found inside the alveolar cells of lung tissues in all the cotton rats immunized with rMuVs (Fig. 8) and the viral antigens inside the alveolar cells in cotton rats vaccinated with rMuV-S79-D1917A were fewest compared to others. A small amount of inflammatory cell infiltration could be observed around the antigen-positive cells, which may be related to virus clearance. No viral antigen was detected in brain tissues from all groups as shown in Supplementary Fig. S5B.
Figure 8. Immunohistochemical (IHC) staining of lungs of cotton rats vaccinated with rMuVs followed by MuV challenge. Cotton rats were immunized intranasally with each rMuV mutant. At week 9 post-immunization, cotton rats were challenged with 1 × 107 PFU of a wild type MuV strain and were sacrificed at day 4 post-challenge. The right lung from each cotton rat (N = 45) was fixed with 4% formaldehyde and embedded in paraffin, sectioned at 4 μm and stained with monoclonal antibody against mumps N protein (Abcam, ab9880, 1/500 dilution in PBS) to determine the distribution of viral antigen. Scale bar: 50 μm.
Design of Amino Acid Substitutions in the MTase Catalytic Site and SAM Binding Site of MuV L Protein
Recovery of rMuVs Carrying Mutations in the MTase Region of L Protein
Replication Kinetics of rMuV-S79s
Genetic Stability of Recombinant rMuV-S79 Mutants in Cell Culture
Replication of rMuV-S79 Mutants in Cotton Rats
rMuV-S79 Mutants Provided Complete Protection Against MuV Challenge
-
A live attenuated vaccine (LAV) is one of the most promising vaccines for human paramyxoviruses and pneumoviruses. LAV has many advantages such as lack of enhanced lung diseases, ease of production, economically feasibility and induction of durable and strong immunity. In China, children receive MMR vaccine (measles, mumps and rubella) since 1990s and S79 mumps attenuated vaccine originating from MuV JL strain was used for MMR vaccination (Pang et al. 2018). Although MMR vaccine is highly successful in controlling measles, mumps and rubella, mumps outbreaks in vaccinated populations have been reported in recent years (Cui et al. 2017, 2018; Ma C et al. 2018; Ma R et al. 2018). Therefore, developing a new MuV vaccine candidate with an improved safety and efficacy is still needed.
Traditional methods for preparing live attenuated mumps vaccines are time-consuming, high-costing and difficult to obtain stable mutations (Buynak and Hilleman 1966). The emerging reverse genetics technology allows us to modify the virus protein genes related to safety and immunogenicity and can obtain virus vaccine candidate strains in a short time, which is more controllable and rapid than traditional methods. Vero cell culture-based vaccine development allows producing high-dose vaccine in a more simplified way, surpassing the traditional technique of serial passage in chicken embryos. The traditional virus rescue method is based on T7 RNA polymerase driven by vaccinia virus, which inevitably suffers the problem of helper virus contamination. In this study, we used BHK cells stably expressing T7 RNA polymerase to rescue rMuVs, which prevents the risk of being contaminated by vaccinia virus. This method has been approved in other studies for rescue of human metapneumovirus, bovine respiratory syncytial virus and measles virus (Ma et al. 2014; Sun et al. 2014; Zhang et al. 2014; Cui et al. 2017; Pang et al. 2018; Wang et al. 2018; Zhou et al. 2019b). We have effectively rescued rMuV-S79 without helper vaccinia virus in our previous researches (Zhou et al. 2019a, 2019b).
We aimed to increase the immunogenicity of the MuV vaccine and reduce its side effects by mutating the methyltransferase catalytic site and the SAM binding site of the L protein. We successfully rescued four rMuV-S79s carrying SAM binding site mutations (rMuV-S79-A1814G, rMuV-S79-G1816A, rMuV-S79-G1818A and rMuV-S79-D 1892A) and four mutants in MTase catalytic site (rMuV-S79-K1792A, rMuV-S79-D1917A, rMuV-S79-K1953A and rMuV-S79-E 1990A). Among these recombinant viruses, rMuV-S79-A1814G showed larger plaque size and faster progress of CPE than the parental virus. The SAM binding site in the L proteins of most NNS RNA viruses is an authentic GxGxG…D motif. However, the SAM binding site of L proteins of MuV, NDV and PIV2 is an AxGxG…D motif, in which the first glycine residue is mutated to alanine. When the alanine is mutated back to glycine in SAM binding site of MuV L protein, the resultant recombinant virus can accelerate viral replication. Thus, the parental rMuV-S79 has a naturally occurring mutation in the SAM binding site, which may contribute to the attenuation of rMuV-S79. All other rMuV-S79 mutants were significantly more attenuated than the parental rMuV-S79, which produced smaller plaque, delayed viral replication and delayed CPE progress. Most of rMuV mutants at the SAM binding site tend to affect the size of the virus plaque, while most of mutants at the catalytic site are more aggressive at affecting the virus titer. This demonstrates that mutations to the MTase catalytic site and SAM binding site further attenuated the MuV, thereby enhancing the safety of the vaccine candidates.
Ferrets and monkeys are susceptible to MuV infection (Rubin et al. 1999; Parker et al. 2013), but their models are huge and expensive. For scientific research, the mouse animal model is a better experimental choice. However, mumps cannot be replicated well or cause disease in adult mice (Tsurudome et al. 1987; Cusi et al. 2001) and cannot be detected in the lungs of normal mice. At present, the newborn Lewis rat hydrocephalus model was used to evaluate the neurotoxicity of mumps vaccine by the severity of hydrocephalus by immunization of mumps virus in the brain (Wolinsky et al. 1976; Kristensson et al. 1984). Similar with the Jeryl Lynn mumps vaccine strain, the Chinese "S-79" mumps vaccine strain has been shown low neurotoxicity in monkey animal experiments (Su et al. 2014). So, we did not use the newborn Lewis rat hydrocephalus model for neurotoxicity testing. Some researchers have used IFN-α/β R-/- mice as an infectious model of mumps and found that IFN-α/β R-/- mice can be detected with a virus titer of 1000 PFU/g lung tissue in the lungs after mumps infection (Pickar et al. 2017), which was useful for evaluating the toxic reaction of mumps virus in mice. Recently, cotton rats have been regarded as improved small animal model for human paramyxoviruses and pneumoviruses including studying viral infection and pathogenesis, testing vaccine efficacy and evaluating antiviral drugs (Boukhvalova and Blanco 2013; Green et al. 2013). In our previous research, we found viruses can be detected in the lung of the cotton rat after mumps infection as in IFN-α/β R-/- mice (Zhou et al. 2019b). In this study, we found that neither the parental rMuV-S79 nor wild type MuV strains caused significant histologic lesions in the lungs of unvaccinated cotton rats. However, both MuV strains replicated in the lungs of cotton rats and mumps viral antigens can be detected inside the alveolar cells of cotton rats. These results suggested that cotton rats might be a good model for viral replication and viral antigen detection in the lungs but not lung pathology. In cotton rats, rMuV-S79-D1917A and rMuV-S79-G1818A showed a similar replication capacity in the lungs and lower virus titer was found in rMuV-S79-G1816A and rMuV-S79-E1990A compared to rMuV-S79. rMuV-S79-K1792A, rMuV-S79-D1892A and rMuV-S79-K1953A did not have detectable infectious virus in the lungs. These results demonstrated that most of these rMuV mutants were further attenuated in replication in vivo.
rMuV-S79-G1816A, rMuV-S79-G1818A and rMuV-S79-E1990A groups produced neutralizing antibodies similar to the parent rMuV-S79. Importantly, rMuV-S79-D1917A triggered higher neutralizing antibodies than the parent rMuV-S79 (P < 0.05), with fewest viral antigen detection provided complete protection against wild type MuV infection. This suggested that rMuV-S79-D1917A had a higher immunogenicity than rMuV-S79. These rMuVs will be ideal candidates for mumps vaccines. rMuV-S79-K1792A, rMuV-S79-D1892A and rMuV-S79-K1953A are safer in vitro/vivo than the rMuV-S79 strain, but produces lower neutralizing antibodies, which were not suitable as candidates of mumps vaccine. It has been recognized that mRNA stability and efficient translation are related to G-N-7 methylation, while 2′-O methylation is important for distinguishing between self and non-self mRNA (Zust et al. 2011; Sun et al. 2014). Both the mutations in mRNA methyltransferase catalytic site and the SAM binding site may affect the level of G-N-7 methylation and/or ribose 2′-O methylation of mRNA. The replication kinetics of rMuVs in Hela cells showed that rMuV-S79-K1792A and rMuV-S79-K1892A were pronounced reduced than the parent strain. The 2′-O methylation functions of these two rMuVs may be affected, but it is uncertain whether there is a loss of G-N-7 methylation function. While rMuV-S79-A1814G and rMuV-S79-E1990A may be less affected by interferon and may be more attenuated by G-N-7 methylation. For rMuV-S79-D1917A, which had a higher immunogenicity than rMuV-S79, we think the change of this site may only affect the catalytic activity of 2′-O methylation. The attenuated mechanism of 2′-O methylation virus enhances the sensitivity to IFN antiviral effects (Daffis et al. 2010) and can also promote the expression of interferon (Zust et al. 2011). Interferon can promote cellular immunity and humoral immunity, thereby affecting the expression level of antibodies. Limited by radioactive experimental conditions, we failed to detect the impact of the respective mutations on the G-N-7 or 2′-O methylation of mRNA methyltransferase. The methylation of mRNA methyltransferase of rMuVs needs to be confirmed by further researches.
Adult mice are insensitive to mumps infection, but newborn mice infected with mumps may cause significant lung histology lesion and even death. Immunizing maternal cotton rats as described, then allow these cotton rats to mate, the newborn cotton rats will obtain the IgG theoretically. Therefore, the neonatal cotton rats may be potential animal model to evaluate histopathology and the protection efficiency of the mumps vaccine. In this study, we only tested the B cell immune response of cotton rat mumps and did not evaluate the specific T cell immune response to the live attenuated vaccine. We will complete the evaluation of vaccine immunity in the further development of vaccines. In future research, we will conduct multiple mutations at multiple sites of mRNA methyltransferase to achieve our goal that increase the immunogenicity of the MuV vaccine and reduce its side effects.
Finally, the current mumps epidemic strain in China is type F (Wu et al. 1998) and the S-79 mumps vaccine is type A strain. The F genotype vaccine produced a higher neutralizing antibody titer against the F genotype strain than the A genotype (Jeryl Lynn) vaccine (Zengel et al. 2017). Besides, the attenuated live mumps vaccine of the F genotype has been used to prevent mumps in China (Liang et al. 2010). The method of improving vaccine safety and immunogenicity in this study can also be applied to the attenuated live mumps vaccine of the F genotype. Furthermore, the mumps virus S79 vaccine strain could as a carrier to express antigenic proteins of F genotype mumps viruses for preventing mumps viral infections with the reverse genetics system in our future research.
-
This work was supported by The National Natural Science Foundation for Young Scholars of China (81901679), The Natural Science Foundation for Young Scholars of Zhejiang Province (LQ19H100005) and China Postdoctoral Science Foundation (2019M662076).
-
ZZ and Y-WH designed and guided the experiments in this manuscriptstudy. XH and YW conducted the main work of the experiment, data analysis and article writing. RL and BW provided help with cell experiments, while MZ and DZ provided support from animal technicians. MZ made an article revision. ZZ and Y-WH finalized the article. All authors read and approved the final manuscript.
-
The authors declare that they have no competing interests.
-
This study was approved by the Ethical Committee of Children's Hospital, Zhejiang University School of Medicine (No. 2013119). Written informed consents were obtained from all patients. All animal procedures conformed to the Guide for the Care and Use of Laboratory Animals and were approved by the Zhejiang University Medical Laboratory Animal Care and Use Committee.
















 DownLoad:
DownLoad: