-
Densoviruses (DNVs),pathogens for invertebrates belong to the subfamily Densovirinae of the family Porvoviridae (32, 36) and share many characteristics with the defective and autonomous vertebrate parvoviruses (3, 30). Densoviruses have been isolated from several insect species belonging to different orders,mainly from Lepidoptera (butterflies and moths) and a few from other orders,including Diptera,Orthoptera,Dictyoptera,and Odonata. All known DNVs package complementary,linear single strands of the DNA into se-parate virions,as do several parvoviruses of vertebrates (e.g.,B19 and AAV) (8) ,and replicate autonomously in the nucleus where they produce a typical dense nuclear inclusions (24, 35). DNVs have little sequence identity with the vertebrate parvoviruses (7). Further studies indicate that there are at least three distinct DNV groups,one genus (Densovirus),exemplified by Junonia coenia DNV (JcDNV) (9, 15) ,a second genus (Brevidensovirus) by the Aedes (1, 4) and shrimp (28) DNVs,and a third genus (Iteravirus) by the Casphalia DNV (13) and Bombyx mori DNV type 1 (19). Most DNVs remain unclassified,since few are characterized with respect to their molecular biology. DNVs are autonomously replicating small icosahedral non enveloped particles,20 to 23 nm in diameter,with their capsid consisting of 4 structural polypeptides denoted VP1,VP2,VP3 and VP4 ranging from 41 kDa to over 100 kDa (17, 25, 33, 34).
Restriction maps of several densovirus genomes have been determined and many symmetrical sites were found (9, 15).Observation of linear double stranded monomers or concatemers and single stranded circular monomers with "panhandle" structures (6, 16) suggest the presence of inverted terminal repeat (ITRs). This structure was confirmed by nucleotide sequences of cloned densovirus genomes (9, 19, 31, 33). The ITRs were found to play essential roles in the process of DNA replication,DNA excision from plasmid vectors and integration into the host DNA (2, 27, 29).
The densovirus of Diatraea sacchararalis (DsDNV) was isolated by Meynadier et al (23) from the Guadeloupe sugar cane borer,D. saccharalis (Lepidoptera Pyralidae) which is a major sugar cane borer in Brazil and Guadeloupe. The estimation of loses by this borer is about $ 100 M per year (21, 22). High pathogenicity combined with a limited host range gives some densoviruses potential as effective insecticides (11). Several densoviruses have been used successfully in biological control of pests in the world (5, 11, 14). However,safety concerns remain as previous reports (10) suggested that densoviruses may infect and transform L cells (from mouse). El-far et al. (18) recently demonstrated that neither L nor other vertebrate cells support replication or transcription of densovirus,either after infection or after transfection by using molecular biology tools. Therefore,the DsDNV isolated from this insect host repre-sents a potential biological control agent for the D. saccharalis pests (20). In this paper,we report the pathogenicity of the DsDNV in its host larvae,the characte-rization of the genome including visualization of DNA molecules by electron microscopy (EM.),the restriction map of the viral genome and the comparison with two well known densovirus genomes of JcDNV and GmDNV.
HTML
-
Eggs and infected larvae of D. saccharalis were received from Campinas University of Sao Paulo,Brazil. Healthy larvae were obtained from CIRAD-CA laboratory(Montpellier,France) and maintained for three ge-nerations in our laboratory on Poitout medium (26).
-
D. saccharalis larvae were reared with solid POITOUT medium and the third star larvae were infected with solid medium containing viruses (8 homogenised dead larvae in PBS buffer).1 200 larvae at third star were equally divided into three lots (400 larvae each),two lots of them were infected with DsDNV and the third lot were fed with Poitout medium containing the same amount of PBS as a control. Dead larvae were collected from the solid medium every three days and counted. For virion purification,infected larvae were first homogenized in PBS buffer or Tris-SDS buffer (50 mmol/L Tris-HCl pH 7.8,0.06% SDS) both con-taining 2% ascorbate. After filtration through cheesecloth and clarification at 10 000 g,5 min.,the virions were recovered by centrifugation at 200 000 g for 2 h. The viral pellet was resuspended in 50 mmol/L Tris-HCl (pH 7.8) buffer and homogenised by ultrasonication,then clarified (10 000 g,5min.). The supernatant containing the virus particles was layered onto a 20-76 renografin gradient prepared in Tris buffer and centrifuged to equilibrium at 100 000 g (SW 28,Beckmann rotor) at 8℃ for 12 to 15 h. The virus band present in the lower part of the gradient was collected and dialyzed against TE buffer (10 mmol/L Tris,1 mmol/L EDTA,pH 8.0) for 48 h. Virion purity was confirmed by mea-suring the OD260/OD280 and by visualizing with electron microscopy (EM.).
-
Purified DsDNV particles were conserved in TE buffer (10 mmol/L Tris,1 mmol/L EDTA,pH 8). For DNA extraction,buffer was adjusted to 10 mmol/L Tris,4 mmol/L EDTA,200 mmol/L KCI 5 mg/mL SDS and 200 μg/mL of proteinase K (final concentration) and incubated at 50℃ for 1~2 h or 60℃ for 30 min. The DNA was phenol extracted and precipitated by two vo-lumes of 100% ethanol in the presence of 0.3 mol/L sodium acetate,for 15 min at -70℃ or overnight at -20℃. The DNA pellet was rinsed twice with 70% ethanol and finally resuspended in TE buffer. The purity and concentration of the viral DNA was determined by UV spectrophotometer. DNA molecules was visualized under an EM. The nicked double-stranded replicative form of X 174 RFII DNA was added to each sample as a control for size calibration.
-
The extracted DNA was treated with a number of endonucleases under the conditions specified by the suppliers. Generally,1 to 2 μg of DNA was digested in a final volume of 20 μL containing 1×digestion buffer and 2~5 U/μg of DNA. The mixture was incubated 1~ 2 h at 37℃. Restriction fragments were analysed using 0.8% ~ 1.8% of agarose gels in TEP buffer (90 mmol/L Tris-Phosphate,20 mmol/L EDTA). Small Fragments (150 bp and less) were analysed by polyacrylamide gels electrophoresis (7% ~ 9%). The gel was stained with ethidium bromide and photographed under UV light. The size of restriction fragments was determined by linear regression using DNA MW marker Ⅵ and Ⅶ (Roche) as standards.
1.1. Viral strain and insect host
1.2. Larval infection,pathogenicity test and virus purification
1.3. Viral DNA extraction and visualization by EM
1.4. DNA digestion and gel electrophoresis
-
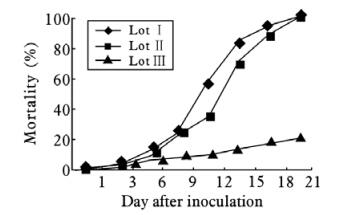
Three lots of D. saccharalis third star larvae were used for this experiment. Two lots (Lot Ⅰ and Lot Ⅱ) were infected with DsDNV while the third Group (GIII) was used as a control. Dead larvae were collected and counted every three days. The cumula-tive percentage of dead larvae from the three lots is presented in Fig. 1. The results showed that up to 4 days after inoculation,cumulative mortality curves were similar for the three groups and the infected larvae started to exhibit the infection symptoms from the forth day. Symptoms of infection of larvae started with anorexia and lethargy followed by flaccidity and inhibition of moulting and metamorphosis. Larvae become paralyzed and stop feeding after 7 days. After 5 days of infection,the cumulative mortality of infected larvae in both infected lots increased significantly and reached 60% for Lot Ⅰ and 40% for Lot Ⅱ after 12 days. Finally,100% of mortality was achieved after 21 days of infection for both Lot Ⅰ and Lot Ⅱ,whereas that of the control group was only 10% and 20%,respectively,after same periods of infection,suggesting that the high mortality of infected larvae groups was due to high pathogenicity of DsDNV. This was confirmed by the purification of DsDNV virions from the infected larvae but not from dead larvae of control group.
-
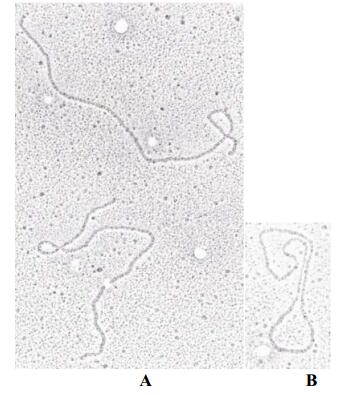
The dsDNA molecules of DsDNV were extracted and examined under the electronic microscope (EM). They appeared in EM as linear molecules (Fig. 2A). Size was measured and calculated ranging from 1.97 to 2.08 μmol/L from 18 viral DNA molecules. The average size of these molecules was estimated to be 2.03 ±0.03 μmol/L. This value was adjusted according to the size of known length of circular dsDNA (replicative form) of X 174 bacteriophage (Fig. 2B). This DNA marker molecule of 3.4×106 MDa was expected to be 1.77 μm in length and appeared to be 1.80 μm in length when observed by EM according to our method. Thus,it is known that 2.95 kb (1.92 MDa) correspond to 1 μmol/L,the dsDNA of DsDNV was calculated to be 3.9 (±0.05) MDa or 5.9 ±0.08kb.
-
The restriction fragments were separated in 0.8% to 1.8% agarose gels. An example of this experiment is shown in Fig. 3. For fragments too small to be visualised on agarose gel,8% ~ and 9 % SDS PAGE was used (see materials and methods). 37 restriction endonucleases were used,only 21 of them cut the viral DNA. Some cleaved once the DNA,others generated from 2 to 9 fragments or more. The number of restriction sites and the size of the generated fragments are summari-zed in Table 1. These sizes were corrected by linear regression using DNA MW marker Ⅵ and Ⅶ (Roche). The restriction enzymes Cla Ⅰ and Pst Ⅰ generated 2 fragments each,while enzymes Asp 700,BamH Ⅰ,EcoR Ⅰ,HindⅡ,Hpa Ⅰ,Nco Ⅰ and Nsi Ⅰ generated 3 fragments. The Bcl Ⅰ,Bgl Ⅱ,Xha Ⅰ and Spe Ⅰ generated 4 fragments; Hae Ⅲ and Hha Ⅰ gave 5 fragments,while Sca Ⅰ gave 6 fragments. Alu Ⅰ,Hinf Ⅰ,Dra Ⅰ,Taq Ⅰ and Sau 3A gave 9 or more fragments each. Due to the limitation of our method,we were not able to detect DNA fragments less than 80 bp. Enzymes that did not cut the viral genome were: Ava Ⅰ,Asp 718,Avi Ⅱ,Bgl Ⅰ,BstE Ⅱ,Dpn Ⅰ,EcoR, HinB Ⅲ,Kpn Ⅰ,Ksp Ⅰ,Pvu Ⅱ,Sac Ⅰ,Sal Ⅰ,Sma Ⅰ,Sph Ⅰ and Xho Ⅰ. The average size of the ds DNA calculated by summing the sizes of the fragments gene-rated by each enzyme,was close to 5.95 kb. The gene-rated fragments were listed from the largest to the smallest (A,B,C,D,E,F) (Table 1). The total length of undigested DsDNV genomic dsDNA was also estimated on 1.2 agarose gel along with digested genomic DNA samples isolated from GmDNV and JcDNV and was densoviruses have similar genome sizes. The sizes of the restriction fragments generated by each enzyme were adjusted according to the value of 5.95 kb estimated for the undigested DNA.

Table 1. Restriction fragments of the genomic DNA of DsDNV.
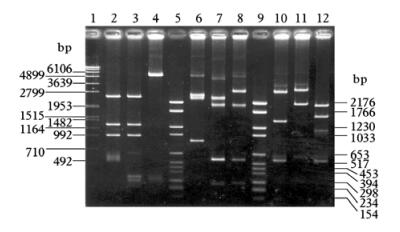
Figure 3. Electrophoretic profile of restricted DsDNV genomic DNA. 1.2% Agarose gel. Lane 1/ 5/ 9,MW marker; Lane 2,Hha Ⅰ; Lane 3,Hha Ⅰ-BamH Ⅰ double digestion; Lane 4,BamH Ⅰ; Lane 6: Eco R Ⅰ; Lane 7,EcoR Ⅰ-Spe Ⅰ double digestion; Lane 8,Spe Ⅰ; Lane 10,Spe Ⅰ-HinC Ⅱ double digestion; Lane 11,Hinc Ⅱ; Lane 12: Nsi Ⅰ-Cla Ⅰ double digestion. Fragments of 80 bp or less were not detectable by this method.
-
The restriction map of the ds DNA of DsDNV was constructed using combination of total and partial digestion with each of the different enzymes,or simultaneous digestion with pairs of restriction enzymes. Since the orientation of the ssDNA of the DsDNV genomic particle is unknown,we arbitrarily decided that from the two fragments generated by Pst Ⅰ digestion,the largest one of 4 300 bp (A) is positioned on the right side (3'end) and the smaller fragment (B) of about 1650 bp is situated on the left side (5'end) (Fig. 4A). All the restriction fragments obtained using the other enzymes were positioned according to the orientation above. A total of 57 restriction fragments were ordered and allowed the construction of a comprehensive restriction map by the positioning of forty one restriction sites (Fig. 4A). The Sca Ⅰ,BamH Ⅰ and Hha Ⅰ restriction sites were situated symmetrically at both extremities,suggesting that there was an ITR structure at both ends.
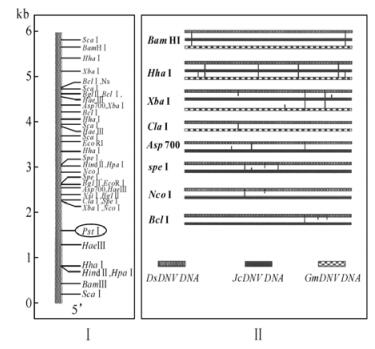
Figure 4. Restriction map of DsDNV genomic DNA (A) and its comparison to that of JcDNV and GmDNV (B).
As mentioned above,GmDNV,JcDNV and DsDNV ds DNA have almost similar sizes of about 6 kb. To iden-tify whether there are some similarities of the restriction map among these DNVs,we compared the restriction maps of the genome of the DsDNV genomic DNA to those of JcDNV and GmDNV (Fig. 4B). The results indicated that the Bam HI restriction sites are similar for all three genomes. The three genomic DNA share four identical Hha Ⅰ restriction sites with 2 additional sites on JcDNV and GmDNV genomic DNA. Xba Ⅰ cleaves twice in similar positions the three genomes,but there is one more restriction site positioned differently on each of the genomes. Cla Ⅰ cleaves DsDNV and GmDNV identically. The Asp 700 restriction sites (2) are similarly positioned on JcDNV genome,even one more restriction site of this enzyme were found on JcDNV genomic DNA. The Spe Ⅰ endonuclease cut three times each genome of DsDNV and JcDNV with two similar positions. Nco Ⅰ and Bcl Ⅰ enzymes have only one restriction site similarly positioned on both DsDNV and JcDNV. Restriction enzymes Alu Ⅰ,Dra Ⅰ,Hinf Ⅰ,Sau 3A,and Taq Ⅰ cut both DsDNV and JcDNV many times. These sites were not mapped on the DsDNV genomes.
2.1. Pathogenicity assay of DsDNV in vivo
2.2. EM visualization of DNA molecules
2.3. Restriction pattern of DsDNV genome
2.4. Restriction map of DsDNV genome
-
Many of the densoviruses isolated so far from different species display outstanding stability (3, 32). Naturally occurring infections by densoviruses in their host larvae in latent forms and the activation of these infections by different stress factors supports the view that these viruses play an important role in controlling and regulating their host populations in nature and hence manifest a substantial potential as a component in integrated pest management. Therefore,densovi-ruses are very attractive agents in biological control (11) ,but are generally not considered,since Kurstak et al (10) reported that a mammalian cell line,L cells from mouse,supports the multiplication of GmDNV. El-Far et al (18) has recently clarified that neither L cells nor other vertebrate cells supports replication or transcription of the densovirus from the Mythimna loreyi (MlDNV) (12) which shares over 90% DNA sequence identity with JcDNV and GmDNV. The DsDNV was isolated in Brazil from the sugar cane borer Diatraea saccharalis which is a major sugar cane pest in Brazil and Guadeloupe (23). Our pathogenicity assay with the DsDNV (Fig. 1) demonstrated that 100% of the tested larvae were killed,underscoring the promise of this virus as biocontrol agent for its natural hosts. Although it takes longer for insect viruses including densoviruses to kill pests than chemical pesticides do,their efficacy can be improved through genetic engineering by inserting an insect-specific toxin gene into these viruses. Jiang et al have recently reported that the efficacy of the smoky-brown cockroach (Periplaneta fuliginosa) densovirus (PfDNV) as a biopesticide was augment-ed significantly by inserting the insectspecific toxin gene BmKIT1 (14).
The genome size of the DsDNV was determined by the electron microscopy visualization of viral DNA molecules and the gel electrophoresis of both native and endonuclease digested DNA fragments. The full length of the DsDNV genomic DNA was estimated to be 5 950 bp ±50 bp by both techniques and is similar to that of the genomes of JcDNV and GmDNV (9, 15, 17). The restriction map of DsDNV was constructed using 16 endonucleases and 41 sites were located. Since the orientation of the DNA molecule was proposed arbitrarily,the 5' and 3' ends have yet to be defined. The Sca Ⅰ,BamH Ⅰ and Hha Ⅰ restriction sites were positioned symmetrically at both extremities,strongly suggesting the presence of inverted terminal repeats (ITRs). The size of the ITR was estimated to be about 500 bp according to the position of Bam HI sites and the homology with the genomes of JcDNV and GmDNV (15, 17). Based on the full length of DsDNV genome of about 6 kb,and possession of a long ITR (arround 500 bp) at both ends,DsDNV could be placed in the same densovirus category (ambisense densovirus,group A) with JcDNV and GmDNV (31).
To determine the level of homologies among these three densoviruses,Southern hybridization experi-ments have also been performed. The probes prepared from each of three densovirues all revealed a very strong reaction to these three viral DNAs (data not shown),suggesting that they share high identities at the level of nucleotide sequences. To obtain further information on the genomic organization,the nucleotide sequences of DsDNV genomic DNA has been cloned and partially sequenced,but 40 ~50 nt at both ends have not yet been determined,the sequenced DsDNV indicated that it shared over 90% identities with JcDNV and GmDNV at both nucleotide and protein levels (data not shown). More detailed studies on pathogenesis and the molecular biology of this virus are needed before it can be used as a biological control agent for controlling the population of Diatraea saccharalis larvae in the field.













 DownLoad:
DownLoad: