-
Coronary heart disease (CHD) is the second highest risk factor for cardiovascular death in China, where the burden of CHD has been increasing (Feng et al., 2016). Recent studies revealed that there may be a link between the gut microbiota and CHD (Wong et al., 2012). Modulation of the gut microbiota is suggested to have the potential to reduce the risk factors associated with CHD (Emoto et al., 2016). Being the main member of the gut microbial ecosystem, gut viruses may also have a potential impact on the microbial ecosystem of the hosts.
Viruses, the most abundant biological entities in the biosphere, have been classified and determined in multiple normal species and ecosystems, such as marine ecosystems, freshwater lakes and the human gut (Wommack and Colwell, 2000; Zhang et al., 2006; Breitbart et al., 2008). However, because of the high diversity of their genomes and morphology, it is hard to cultivate novel viruses or deeply explore their populations. So far, many viruses have not been discovered, and it is difficult to discuss their distribution and composition.
The metagenomic approach, which is a new technology within the last two decades, has provided an in-depth look at the molecular diversity of viruses in a range of environments, including the human gut system (Breitbart et al., 2002; Minot et al., 2011).Microviridae is one of the main families of bacteriophages with ssDNA. They are widely spread in marine environments, freshwater habitats, human gut or feces, stromatolites (Angly et al., 2006; Desnues et al., 2008; Lopez-Bueno et al., 2009; Tucker et al., 2011; Roux et al., 2012a; Roux et al., 2012b) and so on. Based on the nucleotide sequences and the phylogeny of the major capsid protein (VP1), Microviridae are further divided into seven subgroups, including microviruses (genus Microvirus), gokushoviruses (subfamily Gokushovirinae), alphaviruses (subfamily Alphavirinae), the recently described pichoviruses (subfamily Pichovirinae), the new clade Stokavirinae (stoka: small in Sanskrit), Group D and Aravirinae (ara: little in Sanskrit) (Roux et al., 2012b; Quaiser et al., 2015).
In this study, using the viral metagenomics method, we have investigated 43 stool samples from 37 CHD inpatients and six healthy people, and analyzed their virome composition. Twelve divergent Microviridae genomes were determined from the samples, and phylogenetic analysis and genome comparisons were performed. The data from the present study provide new insight of the diversity, distribution and abundance of the Microviridae subfamilies in humans, especially in CHD patients.
-
Thirty-seven fecal samples were collected from CHD inpatients from the Central Hospital in the Minhang District of Shanghai, China, while six fecal samples were collected from healthy residents, without a history of cardiovascular disease, living in communities in the Minhang District of Shanghai, China, as controls. All of the CHD patients had coronary angiographies (CAGs) and percutaneous coronary interventions (PCIs) performed in the cardiovascular department of the hospital, and CHD was defined when the narrowness diameter of the left main coronary artery, the left anterior descending branch, the circumflex artery, the right coronary artery and other main factors were up to or exceeded 50%; while the control members had no history of cardiovascular disease. All the subjects met the following conditions: (1) no application of any antacids, probiotics, antibiotic or antimicrobial agent for the past one or more months; (2) no other digestive system diseases; (3) no surgical operations on their digestive systems; (4) no drunkenness, smoking or diabetes or diseases that might affect the gut microbiota; (5) resident in southern China; (6) aged between 50 and 85 years old.
All samples were preserved at –80 °C. Fecal samples were re-suspended in ten volumes of phosphate-buffered saline and vigorously vortexed for 5 min. Four hundred microliters of supernatant was collected after centrifugation (10 min, 15,000g) and filtered through a 0.45 μm filter (Millipore) to remove eukaryotic and bacterial cell-sized particles. The filtrates enriched in viral particles were treated with a mixture of DNases (Turbo DNase from Ambion, Baseline-ZERO from Epicentre and benzonase from Novagen) and RNase (Fermentas) to digest unprotected nucleic acid at 37 °C for 90 min. The remaining total nucleic acid was then isolated using a QIAamp Mini Viral RNA kit (Qiagen) according to the manufacturer’s protocol. Eight separate pools were randomly generated, five of which contained nucleic acids from five specimens, while three included nucleic acids from six specimens. Eight libraries were then constructed using a Nextera XT DNA Sample Preparation Kit (Illumina) and sequenced using the MiSeq (Illumina) platform with 250 bp paired-end reads with dual barcoding for each pool.
Bioinformatics analysis was performed according to a previous study (Deng et al., 2015). Briefly, paired-end reads of 250 bp generated by MiSeq were debarcoded using vendor software from Illumina. An in-house analysis pipeline running on a 32-node Linux cluster was used to process the data. Clonal reads were removed and low sequencing quality tails were trimmed using Phred quality score ten as the threshold. Adaptors were trimmed using the default parameters of VecScreen, which is NCBI BLASTn (Altschul et al., 1997) with specialized parameters designed for adaptor removal. Human host reads and bacterial reads were subtracted by mapping the reads to human reference genome hg19 and bacterial RefSeq genomes release 66 using bowtie2 (Langmead and Salzberg, 2012). The cleaned reads were de novo assembled by SOAPdenovo2 version r240, using Kmer size 63 with default settings (Luo et al., 2012). The assembled contigs, along with singlets, were aligned to an in-house viral proteome database using BLASTx with an E-value cutoff of < 10–5. The significant hits to virus sequences were then aligned to an in-house non-virus-non-redundant (NVNR) universal proteome database using BLASTx. Hits with a more significant adjusted E-value to NVNR sequences than to virus were removed.
-
Phylogenetic analyses were performed based on predicted amino acid sequences, their best BLASTp matches in GenBank and representative members of related viruses. Sequence alignment was performed using CLUSTAL W (version 2.1) with the default settings. A phylogenetic tree with 1000 bootstrap resamples of the alignment data sets was generated using the maximum-likelihood method based on the Jones-Taylor-Thornton (JTT) model in MEGA7.0. Bootstrap values (based on 1000 replicates) for each node are given. Putative ORFs (open reading frames) in the genome were predicted by the NCBI ORF finder.
-
The genome sequences of SH-CHD 1-SH-CHD 14, respectively, were deposited in GenBank under the accession numbers KX513864, KX513865, KX513866, KX513867, KX513868, KX513869, KX513870, KX513871, KX513872, KX513873, KX513874, KX513875, KX513876 and KX513877.
-
Deep sequencing generated eight raw data sets from eight DNA libraries, including seven experimental groups of CHD patients and one control group. We got 11,273,562 reads from the eight libraries, and 162,319 of them showed significant sequence similarity with known viruses. After genetic optimization, BLASTx searching and classification according to viral taxonomy, the mean size of the assembled contigs was about 1919 bp, and 69±29.6 kinds of viruses were found in each library (Table 1). Using the bioinformatics method, we analyzed the viral community compositions of the eight groups.
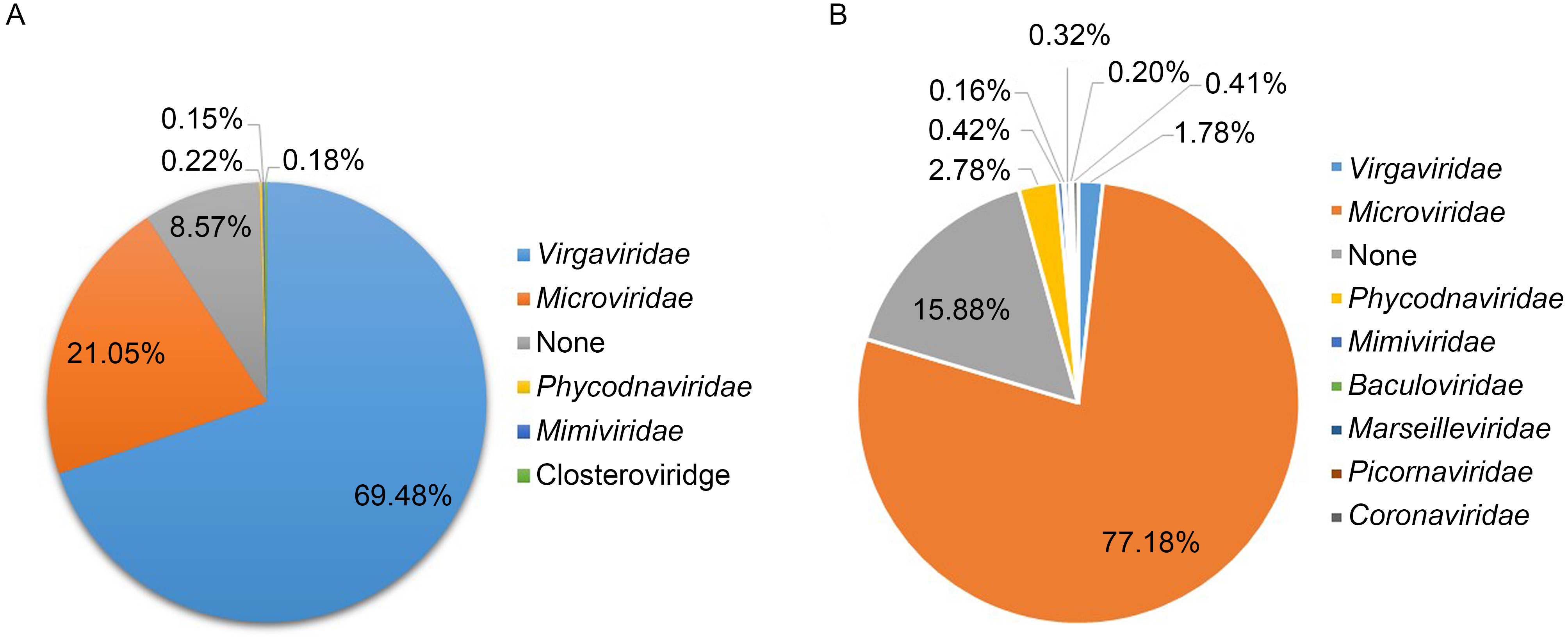
Characteristic CHD group Control group SHu1 SHu2 SHu3 SHu4 SHu5 SHu6 SHu7 Mean percentage SHu8 Total numbers of reads 595670 238206 369360 1555948 2562824 2120470 1560126 – 2270958 Reads of viral origin 3387 474 4553 40624 41025 53041 10362 – 8853 Mean size of assembled contigs (bp) 1715 1816 1623 1811 2569 1537 1421 – 2000 Virus taxa/% ––– ––– ––– ––– ––– ––– ––– ––– ––– Virgaviridae 6.97 24.89 48.74 82.8 31.5 93.9 73.7 69.48 1.78 Microviridae 85.36 67.93 44.85 13.37 41.2 4.97 19.8 21.05 77.18 None 4.19 6.12 5.97 2.82 26.8 0.77 1.23 8.57 15.88 Phycodnaviridae 0.35 0.84 0.09 0.15 0.05 0.05 2.0 0.22 2.78 Mimiviridae 0.26 0.22 0 0.07 0.04 0.08 1.37 0.15 0.42 Betaflexiviridae 1.33 0 0 0.01 0.03 0.002 0.08 0.05 0.002 Closteroviridae 0 0 0.02 0.66 0 0 0.06 0.18 0 Poxviridae 0.37 0 0 0.002 0.01 0.002 0.21 0.03 0.08 Anelloviridae 0.41 0 0 0 0.01 0.002 0.03 0.01 0.10 Picobirnaviridae 0.26 0 0.13 0.01 0.04 0 0.02 0.02 0 Secoviridae 0.09 0 0 0.01 0.002 0.002 0.22 0.02 0.10 Potyviridae 0.06 0 0 0 0.002 0 0.25 0.02 0 Polyomaviridae 0 0 0 0 0 0 0.22 0.01 0 Baculoviridae 0.08 0 0.02 0.01 0.03 0.02 0.03 0.02 0.16 Marseilleviridae 0 0 0.04 0 0.02 0.002 0.13 0.02 0.32 Lipothrixviridae 0.03 0 0 0.002 0.01 0 0.12 0.01 0.05 Astroviridae 0 0 0.02 0 0.01 0.002 0.13 0.00 0 Dicistroviridae 0 0 0 0 0 0.15 0 0.05 0 Asfarviridae 0 0 0 0 0 0.002 0.13 0.02 0 Iridoviridae 0 0 0.02 0.01 0.02 0 0.08 0.01 0.03 Adenoviridae 0.06 0 0 0.002 0.01 0.01 0.04 0.01 0.03 Tymoviridae 0 0 0 0 0 0 0.10 0.01 0 Reoviridae 0.06 0 0 0 0.01 0 0.01 0.01 0 Nudiviridae 0.06 0 0 0 0 0 0.02 0.00 0 Retroviridae 0.03 0 0 0.002 0 0.002 0.03 0.00 0.04 Caulimoviridae 0 0 0.02 0 0 0 0.03 0.00 0 Picornaviridae 0 0 0.04 0 0 0.002 0 0.00 0.20 Alloherpesviridae 0 0 0.04 0 0 0 0 0.00 0.01 Flaviviridae 0.03 0 0 0 0 0.002 0 0.00 0 Togaviridae 0 0 0.02 0 0 0 0 0.00 0 Coronaviridae 0 0 0 0.01 0.01 0 0 0.00 0.40 Arteriviridae 0 0 0 0.01 0.002 0.002 0 0.00 0 Circoviridae 0 0 0 0 0 0 0 0.00 0.04 Note: The dominant species (Microviridae) is marked with italic letters. “None” represents viruses that cannot be identified by BLAST. Table 1. The virus composition of the eight libraries. The plant viruses are marked with bold letters
Primarily, the enteric virome of CHD patients consisted of two dominant families, Virgaviridae (69.48%) and Microviridae (21.05%), while Phycodnaviridae (0.22%), Mimiviridae (0.15%), Closteroviridae (0.18%) and unknown species (8.57%) accounted for the remaining approximately 9%.Betaflexiviridae and Dicistroviridae occupied a portion of about 0.05%~0.1%; a small number of viral reads from Picobirnaviridae, Poxviridae, Anelloviridae, Secoviridae, Potyviridae, Polyomaviridae, Baculoviridae, Asfarviridae, Iridoviridae, Lipothrixviridae, Adenoviridae and Reoviridae were also detected. The numbers of the primarily dominant viral species were different among the different libraries. Three of the seven libraries of CHD patients were dominated by Virgaviridae, while the other four were dominated by Microviridae (Table 1). We calculated the percentage of viral reads of different viral species in each group. Mean percentages were calculated by taking one virus’s total sequences in eight libraries divided by the sum of the eight libraries’ total sequences after BLAST. We listed the mean percentages of viral reads of different viral species (> 0.1%) from the CHD groups and the percentage of viral reads of different viral species (> 0.1%) in the control group to compare the differences in viral composition between them (Figure 1A, 1B). In comparison to the control group, the percentage of enteric viruses in the CHD patient groups was lower; while the percentage of reads of plant viruses, e.g. viruses of Virgaviridae, in the CHD patient group was significantly higher than that in the control group.Virgaviridae is a family of rod-shaped plant viruses, and the CHD patients had eaten more plant-based foods rather than animal products to decrease the fat content of their bodies and keep healthy (Adams et al., 2009; Martinez, 2016). The plant viruses in the CHD patient group were mainly Virgaviridae, which exist in a wide range of herbaceous and monocotyledonous and dicotyledonous plant species (Marais et al., 2015; Quaiser et al., 2015; Schroder et al., 2016).
-
In total, 14 complete circular genomes of Microviridae were assembled from the eight libraries, the genome sizes ranged from 4500 bp to 6400 bp. Genome analysis indicated that all of these complete genomes contained two major ORFs: VP1, encoding the major capsid protein; and VP4, encoding the replication initiation protein. Another Microviridae core gene (encoding minor spike or pilot protein VP2) was detected in all assembled Microviridae genomes except SH-CHD 8.
An internal separation divided the newly assembled Gokushovirinae viruses into two subgroups (group 1 and group 2) (Figure 2). The separation is consistent with the phylogenetic information, expect for SH-CHD 2, which had the same gene order conservation with group 1 but was in group 2. Genomes of Gokushovirinae in group 1 shared the same gene content and gene order (genes encoding VP4, VP5 - DNA binding protein, VP3 - internal scaffolding protein, VP1 and other proteins). All assembled Gokushovirinae genomes in group 2 displayed a reduced content of group 1 conserved genes, with only three genes present (those for proteins VP1, VP2 and VP4), except SH-CHD 2 and SH-CHD 10. SH-CHD 5, SH-CHD 14, SH-CHD 13 and SH-CHD 1 contained an ORF encoding peptidase M15_3.
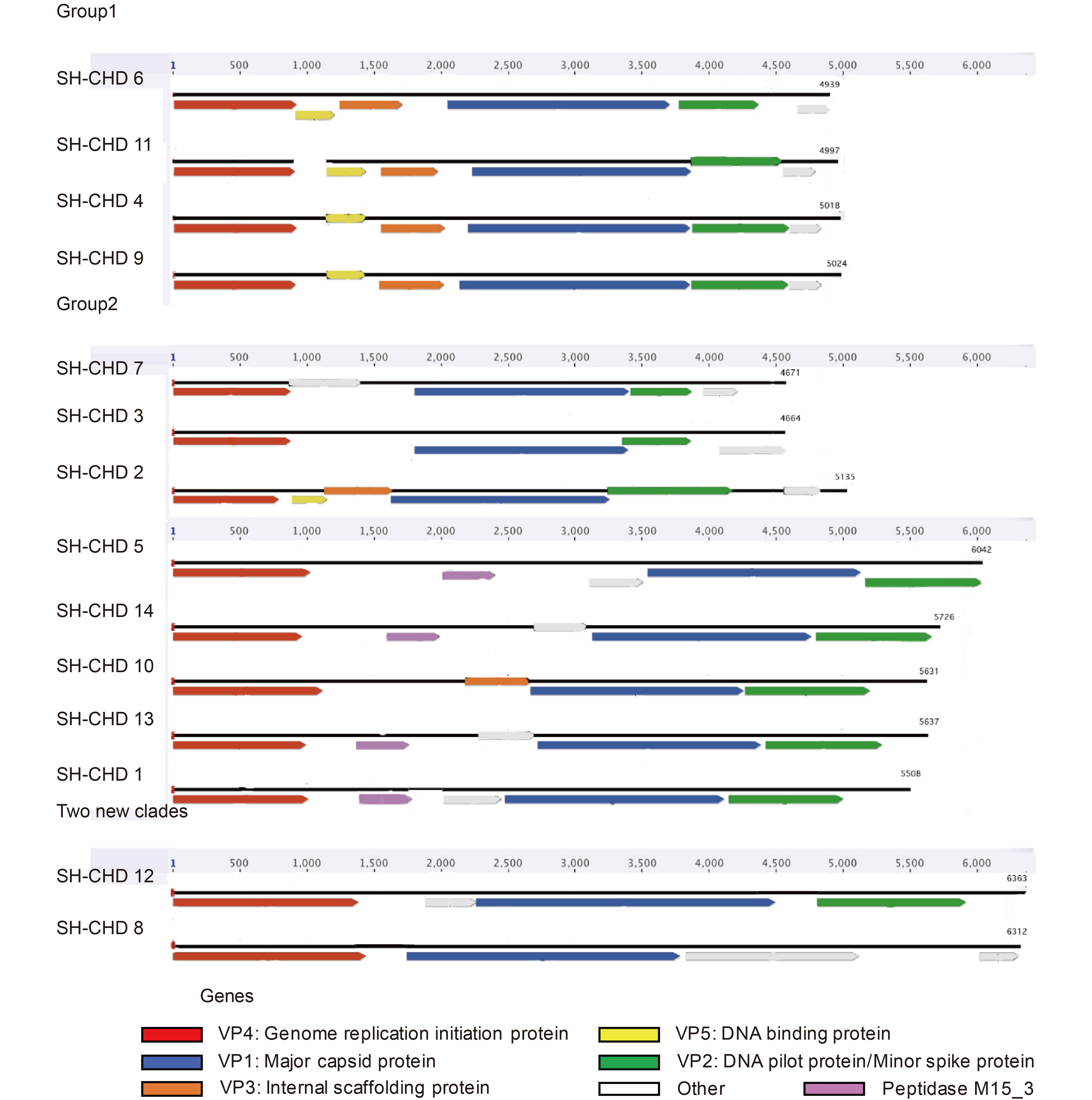
Figure 2. Genome organization of the 14 newly assembled viruses. Linearized genomes are represented for each virus. The ORFs in each genome are colored (VP1, major capsid protein, blue; VP2, DNA pilot protein, green; VP3, internal scaffolding protein, orange; VP4, genome replication initiation protein, red; VP5, DNA binding protein, yellow). Undefined ORFs are colored gray. Peptidase M15_3 is colored purple. The division is consistent with phylogenetic analysis.
Two new clades, SH-CHD 8 and SH-CHD 12, did not have VP3 and VP5 in their genomes. SH-CHD 8 lacked VP2, but possessed a similarly-sized ORF at a position equivalent to that occupied by VP2 in all other members of the Gokushovirinae. The mean genome sizes were bigger than for the other assembled viruses in the present study. The sizes of both VP4 (2018 bp on average) and VP5 (1406 bp on average) of SH-CHD 8 and SH-CHD 12 were longer than for the other assembled Gokushovirinae genomes (1648 bp and 944 bp on average, respectively).
-
In order to explore the diversity and the putative evolutionary origin of the microviruses identified in the present study, a phylogenetic tree was established based on the major capsid protein sequence (VP1). The phylogenetic tree included representative Microviridae subfamilies as references and our 14 newly assembled viruses (Figure 3). Eight well-supported clades were formed: the Microvirus out-group, Group D, Gokusho virinae, Pichovirinae, Aravirinae, Stokavirinae, Microvirus, Alpavirinae and the two strains identified in this study (SH-CHD 12 and SH-CHD 8) (Desnues et al., 2008; Deng et al., 2015).
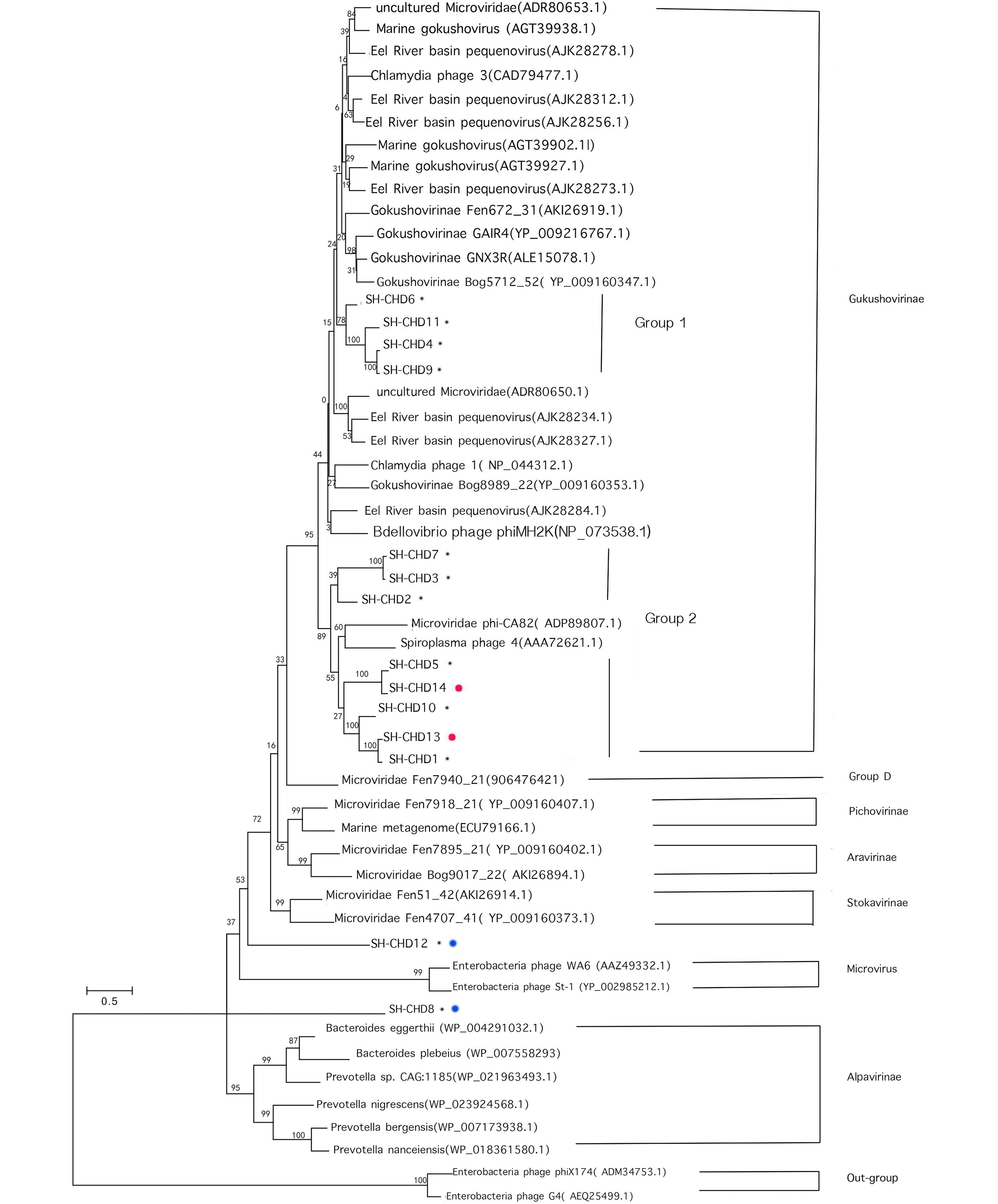
Figure 3. Maximum-likelihood phylogenetic analysis of the major capsid protein (VP1) of the 14 assembled viruses and other environmental microviridae genomes for reference. Black spots indicate viruses assembled from CHD groups. Red spots indicate viruses assembled from feces samples of the healthy control group. Blue spots indicate viruses formed new clades.
Twelve of the newly assembled Microviridae genomes in the present study were grouped into the Gokushovirinae clade, which included another 22 already known representative Gokushovirinae genomes. Ten of these twelve new gokushoviruses were from samples from the CHD groups, and the other two viruses were assembled from the control group. The 12 gokushoviruses identified in this study could be further divided into two subgroups: group1, including SH-CHD 6, SH-CHD 9, SH-CHD 11 and SH-CHD 4; group 2, including SH-CHD 7, SH-CHD 3, SH-CHD 2, SH-CHD 5, SH-CHD 14, SH-CHD 10, SH-CHD 13 and SH-CHD 1. Gokushoviruses in group 1 were most closely related to Bdellovibrio phage phiMH2K (NP_073538.1), while the other group were genetically close to Microviridae phi-CA82 (ADP89807.1) and Spiroplasma phage 4 (AAA72621.1) (Figure 3).
The two new clades (indicated by red spots in Figure 3), represented by SH-CHD 12 and SH-CHD 8, are further separated from other assembled Gokushovirinae. SH-SHD 12 is predicted to be most close to the members of genus Stokavirinae, while SH-CHD 8 falls between genus Microvirus and Alpavirinae. These two genomes of viruses of the family Microviridae may represent two totally new subfamilies or a genus within Microviridae.
-
In the present study, we investigated the enteric virome of CHD patients and healthy controls, which showed significant differences in composition, with slight individual variety among the different CHD groups. In total, the percentage of reads of plant viruses in CHD patients is much higher than that in the healthy control group, while the percentage of reads of enteric viruses in CHD patients is lower than that in the control group. Some research had reported the relationship between the diet and plant viruses. Phan et al. noted the presence of plant viruses, from families such as Virgaviridae, in the virome of the feces of rodents, concluding that these viruses reflected the diet of the rodents; usually plant viruses are considered incapable of infecting humans (Phan et al., 2011). The CHD patients had eaten more plant-based foods, so as to form better eating habits and keep healthy (Martinez, 2016). The viruses in the CHD patients may have suffered effects from various treatments, such as pharmacological treatment applied after CHD (non-)invasive cardiac procedures (PCI, CAG and revascularization) and medicinal inhibition during rehabilitation (Schroder et al., 2016).
In the virome of CHD patients and healthy controls, divergent viral sequences of family Microviridae were found. Fourteen complete genomes of Microviridae were then assembled and analyzed, among which twelve genomes belonged to Gokushovirinae, while the other two genomes may represent two totally new subfamilies or a genus within Microviridae. Our data confirmed and complemented the division of the family Microviridae, as they had the same genome organizational order (VP1 genes, VP2 genes, VP4 genes, VP5 genes (partial) and VP3 genes (partial)) with viruses assembled from human gut (fecal) samples and different from Gokushovirinae viruses assembled from marine (fresh) water samples and chlamydia phages with a gene order of VP1, VP2, VP3, VP4 and VP5 (Roux et al., 2012b; Brentlinger et al., 2002). All the newly assembled Gokushovirinae virus sequences shared the same origin.
Research has found peptidase M15_3 genes in human gut or feces samples have been horizontally acquired by several members of the Microviridae on multiple occasions, and genes transferred between Microviridae are rare (Roux et al., 2012b). SH-CHD 13 and SH-CHD 14 with the peptidase M15_3 genes are viruses assembled from the feces samples of healthy people. It is strange that among 12 newly assembled viruses from CHD groups, only SH-CHD 5 and SH-CHD 1 involved the peptidase M15 genes. Given this, those viruses (SH-CHD 13 and SH-CHD 14) should be assigned to the same clade and separated from the same origin, which is exactly consistent with the phylogenetic analysis result.
The discovery of the two new clades also confirmed the high abundance of Microviridae. The consistent type of difference in their genomes (VP1 genes and VP4 genes) showed that they were probably diverged from one common Microviridae ancestor a long time ago. SH-CHD 8, the only virus without VP2 genes, may be a new clade of Microviridae that separated long ago.
Studies on the gut virome using methods for purifying virus-like particles declared that the normal human gut virome composition was mainly phages of families Siphoviridae, Podoviridae and Myoviridae, followed by members of the family Microviridae (Breitbart et al., 2003; Scarpellini et al., 2015). However, in our CHD groups, the families Microviridae and Virgaviridae were the two dominant ones, while Siphoviridae, Podoviridae and Myoviridae only account for a small percentage of the total (Table 2).Microviridae were previously considered to be exclusively lytic phages, but which can, in fact, integrate into bacterial hosts e.g.Bacteroides and Prevotella spp. in an environment that encourages a temperate (lysogenic) virus-host lifestyle, suggesting that Microviridae could be an important viral family in the human gut (Kim et al., 2011; Reyes et al., 2012). However, the deeper interaction between the human gut virome and CHD still needs to be further explored. Research on both the RNA virome and the DNA viral community revealed that among plant viruses, a pepper-associated virus (pepper mild mottle virus (PMMV)) constituted more than 80% of the identifiable gut viruses (Zhang et al., 2006; Minot et al., 2011; Reyes et al., 2012). However, we did not detect PMMV in the CHD patients in the present study. As plant viruses have a close relationship with food intake and the intestinal bacterial qualitative/quantitative composition of human hosts (Scarpellini et al., 2015), the difference of the plant viruses could be explained by the hosts’ gut microbiota.
Virus taxa Mean composition(%) Virgaviridae 68.46 Microviridae 20.74 Other viruses 8.45 None 2.31 Podoviridae 0.01 Siphoviridae 0.04 Myoviridae 0.00 Table 2. Mean viral composition of the seven CHD groups including major phages
In this study, we have analyzed the composition of the gut virome in CHD patients and healthy controls. Compared with the viral composition of healthy people, viruses of the family Virgaviridae were significantly higher in number in the CHD patient group. The reduction in the number of viruses of the family Microviridae in the CHD groups may be because of the illness of the CHD patients, as Microviridae could be an important viral family in the healthy human gut (Kim et al., 2011). However, we could not find a clear or direct relationship between the gut virome and CHD. We have presented 14 new assembled Microviridae viruses, 12 of them belonging to the subfamily Gokushovirinae, and the other two new viruses presenting two novel Microviridae subfamilies with special gene orders. These new virus sequences expand the currently known genome information, adding to our knowledge of the diversity and distribution of the Microviridae subfamilies.
-
This work was financially supported by the "2014 Agri-X" Project Found of Shanghai Jiao Tong University(No. AF1500028/001).
-
The authors declare that they have no conflict of interest. Ethical Approval was given by Ethics Committee of Shanghai Jiaotong University and the reference number is No. SJTU2015085. An informed consent was obtained from all participants. Consent was obtained from all patients for which identifying information is included in this article.
-
HX CL and ZW designed the experiments. GL, LZ, SQ, YS and LJ carried out the experiments. GL, CL and ZW wrote the paper. All authors read and approved the final manuscript.
Viral metagenomics analysis of feces from coronary heart disease patients reveals the genetic diversity of the Microviridae
- Received Date: 21 October 2016
- Accepted Date: 22 March 2017
- Published Date: 17 April 2017
Abstract: Recent studies have declared that members of the ssDNA virus family Microviridae play an important role in multiple environments,as they have been found taking a dominant position in the human gut.The aim of this study was to analyze the overall composition of the gut virome in coronary heart disease (CHD) patients,and try to discover the potential link between the human gut virome and CHD.Viral metagenomics methods were performed to detect the viral sequences in fecal samples collected from CHD inpatients and healthy persons as controls.We present the analysis of the virome composition in these CHD patients and controls.Our data shows that the virome composition may be linked to daily living habits and the medical therapy of CHD. Virgaviridae and Microviridae were the two dominant types of viruses found in the enteric virome of CHD patients.Fourteen divergent viruses belonging to the family Microviridae were found,twelve of which were grouped into the subfamily Gokushovirinae,while the remaining two strains might represent two new subfamilies within Microviridae,according to the phylogenetic analysis.In addition,the genomic organization of these viruses has been characterized.














 DownLoad:
DownLoad: