HTML
-
Zika virus (ZIKV) is an emerging mosquito-borne flavivirus that belongs to the Flavivirus genus of the Flaviviridae family (Di Guardo 2018). Although ZIKV is mainly transmitted by Aedes mosquitoes, probabl transmission via sexual, perinatal, or transfusion procedures has also been reported (Guedes et al. 2017). While majority of ZIKV infections are asymptomatic, they can cause a wide range of clinical symptoms, such as rash, moderate fever, conjunctivitis, and arthralgia. ZIKV infection has been shown to cause severe neurological complications, such as Guillain-Barrésyndrome (GBS) and myelitis (Petersen et al. 2016). The most serious consequence of ZIKV infection includes microcephaly and other neurological manifestations in newborns of infected mothers (Wiratsudakul et al. 2018). An outbreak of ZIKV infection had occurred in Brazil in which more than 4700 cases of suspected microcephaly were recorded between mid-2015 and January-end 2016. The World Health Organization had declared it a public health emergency of international concern (de Oliveira et al. 2017; Petersen et al. 2016). Since the first imported case reported in China, more suspected ZIKV infections have been recorded, although not confirmed by laboratory methods (Zhong et al. 2016). In China, infections from several co-circulating flaviviruses, such as dengue virus (DENV), occur that are difficult to distinguish from ZIKV infection (Gao et al. 2018). Therefore, a highly specific and sensitive diagnostic method for ZIKV is urgently needed.
ZIKV is a single-stranded RNA virus. Its positive-sense RNA genome can be translated into three structural proteins (C, PrM, and E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (Rastogi et al. 2016). There is a high level of structural similarity between the E protein of ZIKV and those of other flaviviruses, such as West Nile virus (WNV), yellow fever virus (YFV), and in particular dengue virus (DENV) (Dejnirattisai et al. 2016). Nonstructural protein 1 (NS1) is necessary for viral replication and late infection, but is also involved in immune evasion and pathogenesis (Rastogi et al. 2016).
Due to the public health threat of ZIKV infection, multiple diagnostic and detection assays are commercially available at present; however, sensitive and specific diagnosis of ZIKV infections remains a challenge. Traditional polymerase chain reaction (PCR) methods have their limitations, since ZIKV RNA is only detectable in the serum up to 7 days after the onset of symptoms (Reusken et al. 2016). Although plaque-reduction neutralization tests (PRNTs) are highly specific for detecting virus-specific neutralizing antibodies, they are complicated, time-consuming, and not suitable for testing large numbers of samples (Corbett et al. 2015). In contrast, serological assays are rapid, scalable, and technically mature.
NS1 is an important protein that participates in ZIKV replication and virus-host interactions. More importantly, it is a soluble antigen secreted into the bloodstream by cells infected with ZIKV and can induce the production of Abs in the host. The ZIKV NS1 was identified to be largely specific to the virus, and hence, used for the development of anti-ZIKV antibody (Steinhagen et al. 2016). Recently, an enzyme-linked immunosorbent assay (ELISA) based on ZIKV NS1 antigen has been developed and commercialized (Lustig et al. 2017; Steinhagen et al. 2016). Despite the high specificities of this ELISA for anti-ZIKV IgG detection, its main disadvantages include low sensitivities and need of species-specific labelled antibody for non-human samples. In this study, we adopted a new platform named luciferase immunosorbent assay (LISA) for the ultrasensitive detection of ZIKV infection using ZIKV NS1 protein as antigen. The diagnostic performance of ZIKV LISA was evaluated using serum samples from ZIKV-infected patients and animals, and potentially crossreactive sera from patients infected with DENV, Japanese encephalitis virus (JEV), and hepatitis C virus (HCV), along with suitable control samples.
-
Several panels of samples (serum, urine, and saliva) were collected from humans and animals infected with ZIKV (humans, n = 16; monkeys, n = 3; rabbits, n = 3; mice, n = 4), DENV (n = 47), JEV (n = 6), HCV (n = 10) or ZIKV-negative healthy blood donors (humans, n = 40; and 3 animals, 1 for each species). All 16 ZIKV-positive samples, including 13 blood samples, 2 urine samples, and 1 saliva sample were collected from Chinese travelers returning from ZIKV-epidemic areas, and some were confirmed by reverse transcription PCR (RT-PCR) and micro-neutralization (MN) assays. Sampling time covered acute phase and early convalescent stage (≤ 100 days after symptom onset) of ZIKV-infected individuals. Some samples were collected for over a year post-infection. DENV samples (n = 47) were provided by Guangdong Provincial Center for Disease Control and Prevention. JEV and HCV samples were convalescent sera collected from Chinese patients (Table 1). All samples were stored at -20 ℃ or -80 ℃ until further use.
Group Reference No. Source ZIKV-RT-PCR MN ELISA LISA-Full-NS1 LISA-C-NS1 Region Notes Control 1-40 China Serum from blood donors - - - - - Samples from ZIKV patients (n=63) 41 Cambodia Serum; 14 days; acute phase + + + + + 42 Cambodia Serum; 285 days; from #41 patient; - + + + + 43 Venezuela Serum; 10 days; acute + + + + + 44 Venezuela Serum; 3 days; acute phase + - - - - 45 Venezuela Serum; 18 days; from #44 patient; + + - + + 46-53 CHs from SA Serum; from 8 confirmed ZIKV patients + + + + + 54 CH from SA Urine; acute phase + - - - - 55 CH from SA Urine; acute phase + - - - - 56 CH from SA Saliva; from #55 patient; acute phase + - - - - Animal serum (n=13) 57 China Monkey w/o ZIKV-infection - - ND - - 58-60 China ZIKV-infected monkey - + ND + + 61 China Mice w/o ZIKV-infection - - ND - - 62-65 China ZIKV-infected mice - + ND + + 66 China Rabbit w/o ZIKV-infection - - ND - - 67-69 China ZIKV-infected rabbit - + ND + + DENV serum (n=47) 70-106 China anti-DENV IgG (+) - - - - - 107 China anti-DENV IgG (+) - - + + + 108 China anti-DENV IgG (+) - - + + + 109 China anti-DENV IgG (+) - - + + + 110-115 Africa anti-DENV IgG (+) - - - - - 116 Africa anti-DENV IgG (+) - - + + + JEV serum (n=6) 136-141 China anti JEV IgG (+) - - - - - HCV serum (n=10) 142-151 China anti HCV IgG (+) - - - - - +, positive; -, negative; CH, Chinese case; SA, South American; ND, not detected. Table 1. Summary of sample detection in this study.
-
The X-ray structure of ZIKV NS1 (1–352 aa) dimer indicates three functionally distinguishable domains, i.e. a hydrophobic b-roll in a stretch of 1–29 amino acids (aa), a/b Wing domain resembling RIG-I-like fold (38–171 aa residues) and a central b-ladder (C terminal residues of 172–352 aa). In this study, the full-length NS1 coding sequence (1–352 aa) of ZIKV (ZIKV-SMGC-1 strain), N-terminal NS1 (1–172 aa), C-terminal NS1 (172–352 aa), C1-NS1 (172–241 aa), C2-NS1 (230–300 aa), and C3-NS1 (283–352 aa) were amplified by RT-PCR and cloned into the secreted luciferase expression vector pNLF-1-N (Promega, USA) (Fig. 1). Primers used for NS1 amplification in this study were listed in Supplementary Table S1. The sequences of each plasmid construct were confirmed by DNA sequencing.
HEK 293 T cells were seeded in cell-culture plate at>85% confluence in 100-mm2 dishes and transfected with the above 5 μg plasmid using jetPRIME® transfection reagent (Polyplus, FRA) according to the manufacturer's instructions. 48 h after transfection, cells were lysed on ice for 30 min with cell lysis buffer. The cell lysis was then centrifuged at 10, 000×g for 5 min at 4 ℃ to collect the supernatant. Each NLuc-antigen fusion protein was verified by SDS-PAGE and Western blot (WB) with the rabbit polyclonal antibody against ZIKV NS1 protein prepared in our laboratory. b-actin was used as an internal control. At the same time, the NLuc luciferase activities of recombinant NLuc-antigen fusion protein in the supernatant were detected separately. The confirmed supernatants were harvested and stored at -20 ℃ till use.
-
A schematic representation of NS1-based LISA is shown in Fig. 2A, and was also reported in our earlier publication (Wang et al. 2019).

Figure 2. Schematic of the LISA and its validation based on different recombinant proteins. A Luminometer plates were coated with protein G to capture the total IgG antibodies. The recombinant proteins were allowed to bind the specific antibody, followed by the generation of fluorescence signal with the addition of luciferase substrate. B Serum samples from the patient with ZIKV infection, collected on day 14 (No. 41) and day 285 (No. 42) after admission in the hospital, were serially diluted and determined the relative fluorescence intensity (RFI) use the method as described. Serum from healthy blood donor (No. 40) was used as negative control. Cut-off value was decided based on values from 40 healthy donors (Nos. 1–40). C Five ZIKV infection serum samples and six negative control human sera were tested by five sets of LISA. The dilution ratio of sera was 1:100. D Serum samples from ZIKV-infected rabbit (No. 67) and negative rabbit (No. 66) were serially diluted and dispensed into the wells of a plate, and tested by five sets of LISA. Each sample was tested in duplicate. Data are representative of at least two independent experiments.
Costar 96-well flat-bottomed luminometer plates (Corning Costar, Corning, NY, USA) were coated with Protein G (5 μg/mL, 100 μL/well) in carbonate buffer (pH 9.6) overnight at 4 ℃. After three times washes with PBS containing 0.05% Tween 20 (PBS-T), each well was incubated with a blocking solution consisting of 5% nonfat milk in PBS for 1 h at 37 ℃. Then, the wells were washed four times with PBS-T. 100-μL aliquots of serially diluted sera were added to the wells, followed by further incubation for 1 h at 37 ℃. After four washes with PBS-T, the plates were incubated with 50 μL of dilute NLuc-NS1 fusion protein for 30 min at 37 ℃. After washing, 50 μL of luciferase substrate (Promega, USA) was added to each well. Relative fluorescence intensity (RFI) values were determined using a GloMax luminometer (Promega, USA). All steps were performed according to the manufacturer's instructions. Each sample was tested in duplicate. The average fluorescence value for the negative controls was calculated, and the cut-off value was determined as twice the average of negative controls. Sera from healthy blood donor, mixed in equal volumes, served as negative controls. Data were expressed as mean RFI of parallel duplicate wells and corrected for background by subtracting the RFI value of the well incubated with 293 T cell extracts in absence of sera.
To avoid the difference of transfection efficiency and protein expression of different batches of preparations, we measure the luciferase activity of crude cell lysates to determine the relative fluorescence intensity (RFI), which is usually between 108 and 1011. ZIKV antigen are always added in each reaction with 107 RFI for LISA. In addition, we include positive and negative controls in each reaction plate to keep the results consistent and reproducible.
-
All samples were subjected to ZIKV NS1-based ELISA (Euroimmun, Lübeck, Germany) according to the manufacturer's recommendations (Lustig et al. 2017; Steinhagen et al. 2016).
-
Samples were subjected to micro-neutralization assay in Vero cells (ATCC, Manassas, VA, USA) (Wong et al. 2017; Xu et al. 2018). Diluted samples (1:100) were mixed with virus particles (1:2000 dilution) of 100 TCID50 (50% tissue culture infective dose) ZIKV-SMGC-1 (kindly provided by Dr. Yang Yang, Shenzhen Third People's Hospital, Guangdong Province, China) in a 10% fetal calf serum-supplemented medium and incubated at 37 ℃ for 2 h. The mixtures were then distributed into 96-well plates containing monolayers of Vero cells with 10% fetal calf serum. After 4 days incubation at 37 ℃ in 5% CO2, the cells were washed with PBS and then fixed with methanol and ethanol (1:1 mixed) for 20 min at -20 ℃. After removal of the fixative, each well of the plates was then incubated with blocking solution consisting of 5% non-fat milk in PBS for 30 min at 37 ℃. The wells were washed three times with PBS-T, added with diluted human monoclonal antibody Z6 against Zika (Wang et al. 2016), followed by further incubation for 2 h at 25 ℃. After three times washes with PBS-T, the plates were incubated with horseradish peroxidase (HRP)-labeled goat anti-human immunoglobulin G (IgG, Beijing Zhongshan Jinqiao Biotechnology Co., Ltd., Beijing, China) for 1.5 h at 25 ℃. 3, 3', 5, 5'-tetramethylbenzidine (TMB) (Sigma, St. Louis, Mo, USA) was added (100 μL/well) after four times washes with PBS-T, and the wells were incubated for 5 min at 25 ℃. Then, 50 μL of 2 mol/L H2SO4 was added to each well to terminate the reaction, and the optical density (OD) was immediately read at 450 nm. The value of Vero cells treated with virus alone (reference group) was defined as 100% infection. Cells treated with PBS alone were included as a background control. The results were expressed as the percentage of infection compared with those of the control group. And the cut-off values were determined as 50% infection reductions. All experiments were performed at least three times.
-
All data were analyzed using GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA, USA) and SPSS (IBM, New York, NY, USA). Pearson correlation coefficients between the different assays were calculated, and statistical significance was defined by a 2 value of < 0.05. Comparative analysis of virus sequences was performed by Clustal Ω software, available on www.ebi.ac.uk/Tools/msa/clustalo/.
Samples
Expression of Different NLuc-Antigen Fusion Proteins
Development of LISA Based on Different NS1 Fusion Proteins
ZIKV NS1-Based Enzyme-Linked Immunosorbent Assays (ELISA)
ZIKV Micro-Neutralization Assay
Statistical Analysis
-
To establish a new approach for ZIKV detection, we first constructed six luciferase expression plasmids containing either full-length ZIKV NS1, N-terminal of NS1, C-terminal of NS1, C1-NS1, C2-NS1, or C3-NS1 (Fig. 1A). These plasmids were confirmed by restriction endonuclease reactions and gel electrophoresis. The NLuc-NS1 fusion protein, expressed by mammalian 293 T cells, was detected using the rabbit polyclonal antibody against ZIKV NS1 in Western blot (Fig. 1B). These results confirmed the correct construction and expression of ZIKV NS1 plasmid. The recombinant proteins may be used in ZIKV-LISA.
-
To optimize the antigenic domain for anti-ZIKV IgG detection based on the six recombinant proteins, we established six assays: Full NS1-LISA, N-NS1-LISA, C-NS1-LISA, C1-NS1-LISA, C2-NS1-LISA, and C3-NS1- LISA. All the serum samples from ZIKV-infected cases were readily detected by the Full NS1-LISA and C-NS1 LISA at up to 1:1600 dilution; however, it was not detectable by N-NS1 LISA [Fig. 2B, only present the data of samples collected on day 14 (No. 41) and day 285 (No. 42)], thereby indicating that the binding domain for antiZIKV IgG is located at the C-terminal domain (172aa–352 aa) of NS1 protein.
We further divided the C-terminal part of NS1 into three fragments (C1, C2, and C3, as shown in Fig. 1) to characterize the binding domain for anti-ZIKV IgG using LISA. Although the C terminus of NS1 showed as high sensitivity as full NS1 in LISA (Fig. 1B), the C1, C2, and C3 domains of NS1 antigen were not able to distinguish ZIKV-infected cases and negative control, even in 1:100 dilution of ZIKV-infected human serum (Fig. 2C). We further performed the assays using the ZIKV-infected rabbit serum. Results showed that full NS1-, C-LISA could differentiate positive sera from negative ones even at the dilution of 1:25, 600, while C1-, C2-, C3-LISA could not at the dilution of 1:1600, the latter ones seemed at least 16-fold lower sensitivity than the former ones (Fig. 2D).
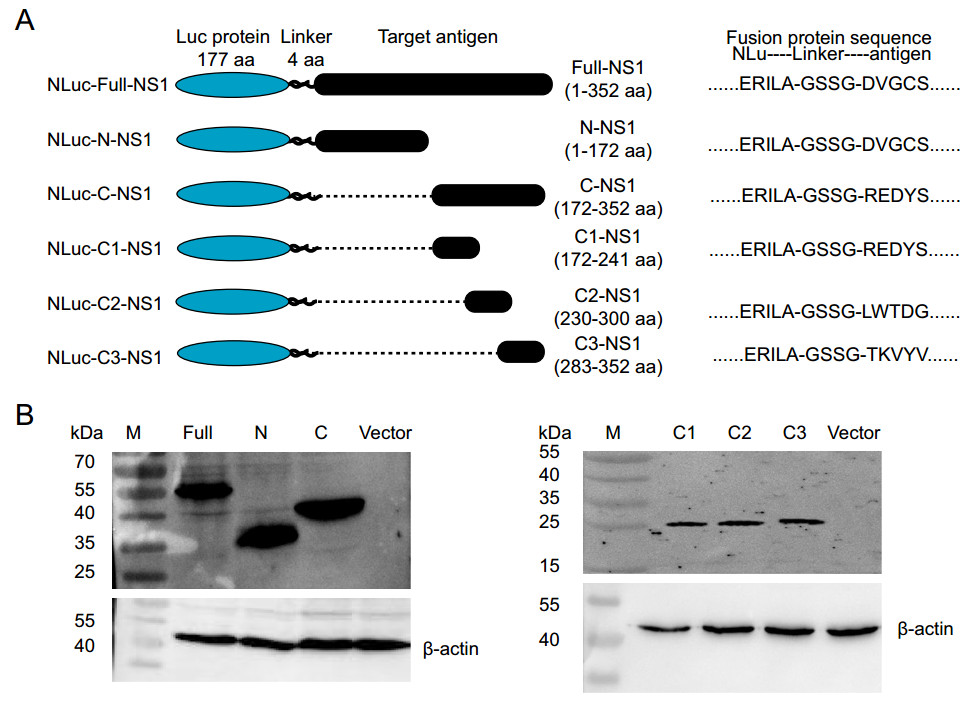
Figure 1. The NLuc-antigen fusion proteins expressed in mammalian 293 T cells. A The structure of six NLuc-antigen fusion proteins. The NS1 fragments were fused to the end of secreted Nano luciferase gene NLu using the "linker" (GSSG), and cloned into the luciferase expression vector pNLF-1-N. B Western blot analysis. Six NLuc-antigen fusion protein, including NLuc-Full-NS1, NLuc-N-NS1, NLuc-C-NS1, NLuc-C1-NS1, NLuc-C2-NS1, and NLuc-C3-NS1 were detected by the rabbit polyclonal antibody against Zika virus NS1 protein.
No reactivity was detected in the samples obtained from 40 healthy blood donors, thus suggesting the high specificity of our assays. Taken together, our results indicate that the C-terminus (172aa–352aa) of NS1 protein may be the optimal antigenic domain for anti-ZIKV IgG detection.
-
We next focused on the full NS1-LISA and C-NS1-LISA to further evaluate their detection ability of anti-ZIKV IgG, relative to ZIKV RT-PCR, commercial NS1-based ELISA, and MN assays (Table 1).
A good correlation between LISA and ELISA was observed with Pearson's correlation coefficients < 0.8 (Fig. 3A). Specificity of ZIKV LISA and ELISA was further validated by MN. The 12 ZIKV-positive serum samples detected by LISA were all positive by ZIKV MN. However, 4 false positive samples (Nos. 44, 54, 55, and 56) were confirmed to be negative by ZIKV MN (Table 1). We also identified a good correlation between MN and LISA (Fig. 3B).
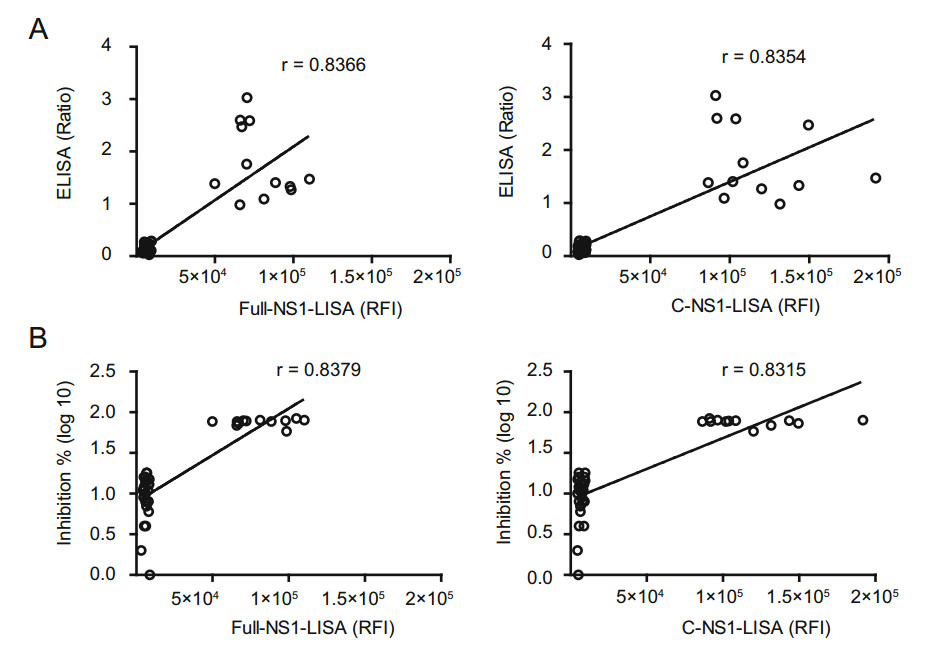
Figure 3. Correlations between luciferase immunosorbent assay (LISA) and enzyme-linked immunosorbent assay (ELISA), and the ZIKV micro-neutralization (MN) assay for detecting anti-ZIKV NS1 IgG antibody. A Correlation between LISA and ELISA based on 12 positive samples and multiple control (negative) samples shown in Table 1. The relative fluorescence intensity (RFI) of the Full-NS1- and C-NS1-based LISA is plotted against the ratio of optical density of ELISA; P < 0.001. B Correlation between LISA and the ZIKV micro-neutralization (MN) assay based on the 12 positive samples and multiple control (negative) samples. The relative fluorescence intensity of the Full-NS1- and C-NS1-based LISA is plotted against the inhibition of neutralization assay; P < 0.001. The neutralizing activity indicate as inhibition % (Inhibition % = (RFI in viral infection well - RFI-sample well)/RFI in viral infection well 9 100%).
In the current study, we included two urine samples (Nos. 54, 55) and one saliva sample (No. 56), both of which were tested ZIKV RNA-positive by RT-PCR. However, neither of LISA and ELISA could detect antiZIKV antibody in these samples (Table 1). In addition, one serum sample (No. 42) collected on day 285 post-infection, was negative for ZIKV RNA by PCR, but anti-ZIKV antibody-positive by both LISA and ELISA, hence indicating the persistence of anti-ZIKV IgG after clearance of ZIKV viremia.
The limit of detection (LOD) of LISA was evaluated in five serum samples from patients with confirmed ZIKV infection, including 4 convalescent patients (Nos. 41–43 and 45) and one acute phase patient (No. 44). Both LISA and ELISA were unable to detect the acute ZIKV infection in the sample (Table 1, Fig. 4). For the four samples from convalescent Zika patients, ELISA was able to detect antibodies till 1:400 dilution, whereas the two LISAs could detect them up to 1:1600 dilution (Fig. 4). According to our results, LISA is at least fourfold more sensitive than the commercial ELISA. We used the twice the average of 40 negative controls as the cut-off for both ELISA and LISA in the Fig. 4, indicating that LISA is superior to ELISA.
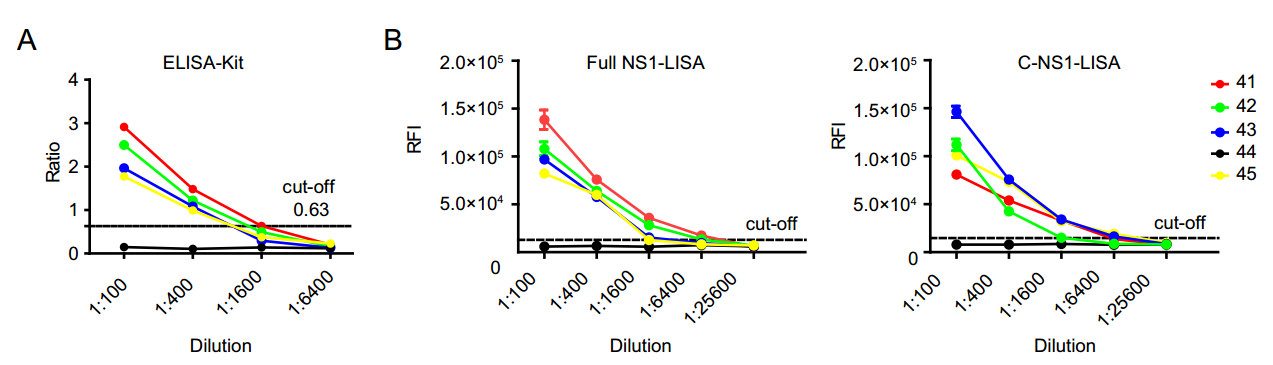
Figure 4. Sensitivity of enzyme-linked immunosorbent assay (ELISA; A) and luciferase immunosorbent assay (LISA; B). Serum samples from 4 convalescent patients (No. 41–43, 45) and one acute phase Zika patient (No. 44) were collected on day 6. Since negative control had very poor response, its results are not shown to avoid interference with the target profiles. Each sample was tested in duplicate. Data are representative of at least two independent experiments.
Among the 13 serum samples from ZIKV-infected patients, both full-NS1-based and C-NS1-based LISA detected the specific IgG in 12 samples while NS1-based ELISA did so for 11 samples. The only serum sample (No. 44) missed by both ZIKV LISA and ELISA was collected on day 3 post-infection, so that means this missed detection is probably due to the lack of anti-ZIKV IgG at such an early stage post-infection. Both LISA and MN could detect a ZIKV RNA-positive sample (No. 45), which was collected on day 18 post-infection and tested negative by ELISA (Table 1).
-
We infected monkeys, mice, and rabbits with ZIKV and detected anti-ZIKV antibodies using the established LISA method. All the ten serum samples from ZIKV-infected animals were tested as positive by full-NS1-LISA (Fig. 5) and were confirmed by ZIKV MN (Table 1). When diluted to 1:1600, the anti-ZIKV antibody was still detectable in the serum samples from ZIKV-infected monkeys and mice (Fig. 5A, 5B). Interestingly, anti-ZIKV titers reached up to 1:25, 600 for ZIKV-infected rabbits (Fig. 5C); no antiZIKV antibody was detected in the uninfected animals (Table 1, Fig. 5). These results clearly indicate that ZIKV LISA could clearly detect the infection in the serum samples from different ZIKV-infected animals, without the need of any species-specific labeled antibody.
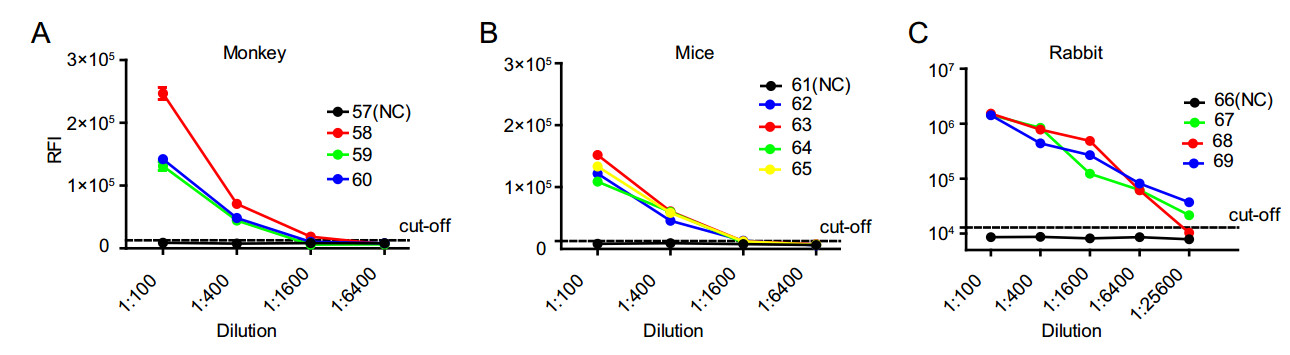
Figure 5. Sensitivity of three different animal serum samples in FullNS1-LISA. Samples from A three ZIKV-infected monkeys (Nos. 58–60) and one negative monkey (No. 57), B four ZIKV-infected mice (Nos. 62–65) and one negative mouse (No. 61), C three ZIKVinfected rabbit polyclonal antibodies (Nos. 67–69), and one irrelevant rabbit polyclonal antibody (No. 66) were tested by Full-NS1-LISA. Each sample was tested in duplicate. Data are representative of at least two independent experiments.
-
Cross-reactivity of ZIKV with DENV, as well as with other flaviviruses has been consistently reported (Balmaseda et al. 2017; Dejnirattisai et al. 2016). We also explored cross-reactivity of ZIKV NS1-based LISA using a total of 63 convalescent sera from people infected with different viruses of the Flaviviridae family, including DENV, JEV, and HCV. We compared the detection results of ZIKV NS1-based LISA and ELISA as well as that of MN (Table 1). False positivity was observed in 8.5% (4/47) of DENV-positive samples for ZIKV NS1-based LISA or ELISA. No cross-reactivity was observed in either of the 6 JEV or 10 HCV-infected sera. To explore the reason of cross-reactivity among the flaviviruses, we aligned their NS1 protein sequences, and found that many members of the Flaviviridae family shared high sequence similarity at the C-terminus of NS1 protein (Fig. 6). The amino acid homology between ZIKV-NS1-C (172–352 aa) and corresponding segments of several viruses (DENV, JEV, WNV, and YFV) is more than 50% (Table 2).
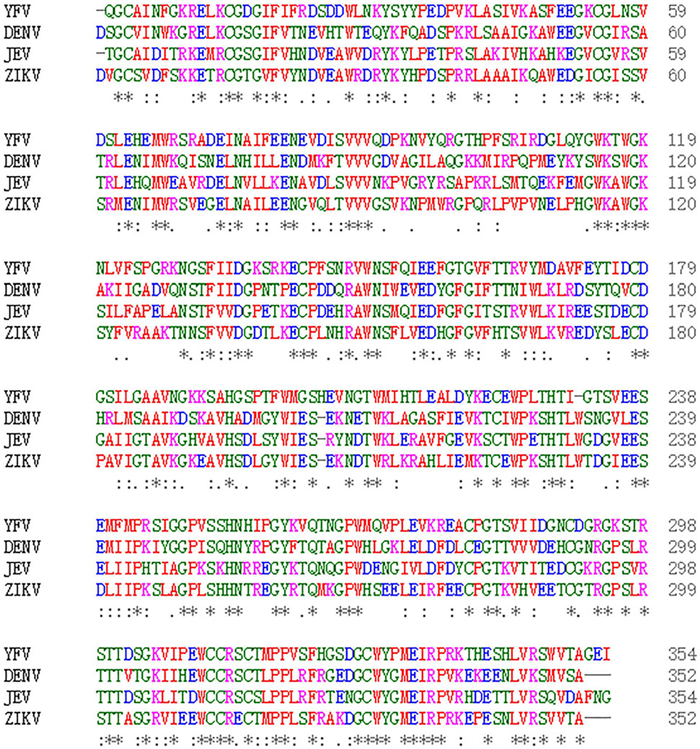
Figure 6. Comparative analysis of four different Flavivirus NS1. Comparative analysis of NS1 was performed by CLUSTAL X software, available on www.ebi.ac.uk/Tools/msa/clustalo/. NCBI accession numbers for the different viruses are: ZIKV (BeH819015), DENV (NP_059433.1), YFV (NP_041726.1), and JEV (AAA81554.1). The conserved sequences are shown by asterisk (*), amino acids that are strongly similar in their properties are indicated by colon (:), and gaps among the sequences are shown by dashes (–). The color codes represent the different types of amino acids.
Virus ZIKV-NS1 ZIKV-NS1-C(aa: 172-352) ZIKV-NS1 100 100 DENV-NS1 54 61 JEV-NS1 56 61 WNV-NS1 56 60 YFV-NS1 47 51 HCV-NS1 44 42 NCBI accession numbers for the different viruses are: ZIKV (BeH819015), DENV (NP_059433.1), JEV (AAA81554.1), WNV (ADZ13193.1), YFV (NP_041726.1), and HCV (BAJ07247.1). Table 2. Comparison of amino acid homology between ZIKV NS1 and corresponding segments of different Flaviviridae members.
Expressionof Recombinant Proteinswith Luciferase Fused with Various Fragments of ZIKV NS1 Protein
The Optimal Antigenic Domain for Anti-ZIKV IgG Detection
Comparison of the Novel LISA with Commercial ELISA and MN for the Detection of Anti-ZIKV IgG
Application of LISA in the Detection of Anti-ZIKV IgG in Various Animal Hosts
Cross-Reactivity of ZIKV NS1-Based LISA
-
ZIKV is an emerging infectious disease that has caused several epidemics worldwide (Franca et al. 2016; Johansson et al. 2016; Moura da Silva et al. 2016). Like other flavivirus infection, both E and NS1 proteins play an important role in ZIKV infection (Yu et al. 2017). Unlike the extensive cross-reactivity between antibodies and E proteins in flaviviruses, NS1 protein seems to be more specific. Antibodies to nonstructural protein 1 (NS1) were largely ZIKV-specific and hence used for the development of a serological diagnostic tool (Franca et al. 2016; Johansson et al. 2016; Moura da Silva et al. 2016). Since emerging or re-emerging infectious diseases are becoming increasingly common, and most of them are from animals, it is very important to urgently establish antibody detection methods for new pathogenic infection and conduct traceable investigation of animal epidemic foci. Here, we present the first report of a novel LISA assay that is potentially useful for rapid and sensitive detection of anti-ZIKV IgG in both humans and animals.
In the current study, we used a new luciferase, NLu, and ZIKV NS1 protein to establish a novel and ultrasensitive approach named LISA for detecting ZIKV infection. NLu can generate a glow-type luminescence (signal halflife < 2 h) with 150-fold greater activity than renilla luciferase, which is commonly used in a luciferase-immunoprecipitation system (Ludolfs et al. 2009). Our results illustrated that both full-length NS1 protein and C-terminal portion of NS1 protein of ZIKV were highly efficient in detecting ZIKV infections in both ZIKV-infected humans and animals. Our results were consistent with those reported by Lee et al., which identified eight epitopes in ZIKV NS1 protein, most of them located in the C-terminus of NS1 (Lee et al. 2018). Furthermore, we identified that the optimal antigenic domain for anti-ZIKV IgG detection was located within 172–352 amino acids (aa) of NS1 protein.
Performance of the only commercial ZIKV NS1-based ELISA has been assessed by different groups (Steinhagen et al. 2016). We compared the results of our LISA with those of the commercially available NS1-based ELISA kit, which has been validated and applied in many studies (Lustig et al. 2017; Steinhagen et al. 2016). The LISA results showed excellent correlation with those of ELISA (P < 0.001) and neutralization assay. Moreover, we found that LISA was able to detect much lower LOD of antiZIKV antibody than ELISA.
LISA has the advantages of rapid development, ultrasensitivity, high throughput, and easy operation. While traditional serological methods need purified antigens for detection, and the purification process is usually very complicated and time-consuming, LISA can directly use cell lysates for detection without the need of purification and post-expression modification. In addition, traditional ELISA method requires enzyme-labeled antibody of the corresponding species, which are not necessary in LISA. Since it is difficult to obtain genus-specific antibodies from vectors such as mites, mosquitoes, and other non-laboratory animals, such as bats, the scope of application of ELISA is limited. Notably, LISA does not need speciesspecific labeled secondary antibodies for detection, which makes it universally applicable for tracing samples from various animal or vectors.
In a recently published report, NS1-based capture ELISA was used to investigate the cross-reactivity of NS1 proteins using convalescent sera obtained from DENV- or ZIKV-infected patients (Gao et al. 2018). Approximately 36%–40% plasma samples from DENV infection donors, including secondary DENV infection donors, showed cross-reactivity with ZIKV (Yu et al. 2017). In the current study, we also tested panels of samples from patients infected with different viruses including DENV, JEV and HCV. The cross-reactivity of ZIKV NS1-based LISA existed in about 8.47% of the samples infected with DENV but not in the samples with JEV and HCV infections. In future, cross-reactivity of ZIKV NS1-based LISA should be further investigated using more panels of samples from cases infected with other flaviviruses, including yellow fever, West Nile virus (WNV), Chikungunya virus (CHIKV), and Tick-borne encephalitis virus (TBEV). And we plan to focus on identifying the sequences that cause cross-reactivity, and refine our LISA for more specific detection of anti-ZIKV IgG based on optimal antigens (Lee et al. 2018).
In conclusion, we have developed a novel ZIKV NS1- based LISA, for sensitive detection and diagnosis of ZIKV infection. Since the three assays showed strong correlation, LISA may be adopted as an alternative to ELISA and ZIKV neutralization assay for confirmatory detection of ZIKV infection. As a high throughput detection platform, it may be useful for large-scale serological epidemiological studies of ZIKV infection.
-
This work was supported by the following grants: the National Key Research and Development Program of China (2016YFD0500301), the National Major Project for Control and Prevention of Infectious Disease in China (2018ZX10101002 and 2017YFC1200503), the Bureau of Science and Information Technology of Guangzhou Municipality, China (201604020011, 201704020219), and National Key R&D Program of China (2018ZX10732401). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank all the staffs from respective authorities of China for providing all samples.
-
ST and WT conceived and designed experiments. TW, YZ, YD and W Wu performed the experiments. TW, W Wang and WT analyzed the data. DW, ZC, W Wu and W Wang contributed reagents/material/analysis tools. TW, ST and WT wrote the manuscript.
-
The authors declare that they have no conflict of interest.
-
A total of 151 samples from human and animal were used in this study, which was approved by the Ethics Committee of Southern Medical University (SMU No. 20160428) and the National Institute for Viral Disease Control and Prevention of China (IVDC No. 2016072501), as well as the waiver of the Informed Consents.
















 DownLoad:
DownLoad: