HTML
-
The genus Bocaparvovirus (BOV), belonging to the subfamily Parvoviridae of the Parvovirinae, comprises small, non-enveloped, single-stranded DNA viruses. The BOV genome is approximately 5.0 kilobases (kb) and includes three open reading frames (ORFs); NS1, NP1, and VP1/ VP2. NP1 is located between the genes encoding NS1 and VP1/VP2 (Manteufel and Truyen 2008). BOVs can infect a large number of mammals, including humans and non-human primates (NHPs). Human and NHP BOVs are separated into two species. One is Primate bocaparvovirus 1, which includes human bocaparvoviruses 1 and 3 (HBOV1 and HBOV3), gorilla bocaparvovirus (GBOV), and chimpanzee bocaparvovirus (CPZh2) genotypes (Cotmore et al. 2014; Qiu et al. 2017). The other species is Primate bocaparvovirus 2, which includes human bocaparvoviruses 2a–c and 4 (HBOV2a–c, HBOV4) (Cotmore et al. 2014; Qiu et al. 2017). HBOV1 was first detected in 2005 in children with respiratory diseases, after which it was identified in people with respiratory symptoms, especially young individuals, worldwide (Qiu et al. 2017; Allander et al. 2005; Schildgen 2013). HBOV1 is considered to be a primarily respiratory pathogen (Qiu et al. 2017; Schildgen 2013; Kapoor et al. 2010a). HBOV2–4 have been linked primarily to gastrointestinal symptoms, although their pathobiology remains unknown (Qiu et al. 2017; Broccolo et al. 2015).
Since 2010, when the first NHP BOV (GBOV) was identified in stool samples from western captive gorillas with acute enteritis diarrhea and its possible association with gastrointestinal diseases was indicated, a further series of BOVs was detected in NHPs (Kapoor et al. 2010b; Sharp et al. 2010). Brozova et al. reported a novel genotype strain (CPZh2) of primate BOV1, which originated from recombination between HBOV3 and HBOV1/ GBOV, in fecal samples from free chimpanzees in Cameroon (Brozova et al. 2016). However, these viruses were somewhat distinct from HBOVs, with more than 8% nucleotide (nt) sequence divergence (Brozova et al. 2016). Interestingly, Kumakamba et al. identified BOV DNA in blood and tissue samples from African NHPs in the Democratic Republic of Congo, and those strains identified strongly (> 97%) with HBOV2/3, rather than with those in gorillas and chimpanzees (Kumakamba et al. 2018). These findings suggested the cross-species transmission of HBOVs between humans and NHPs. Although a growing number of studies have reported related HBOVs in stool samples from gorillas and chimpanzees, the evolution of HBOVs remains unclear. Thus, the identification of novel BOVs is crucial to further our understanding of the evolution and genetic diversity of BOVs and to enable the prediction of cross-species emergence and transmission.
Here, we report on the discovery of a novel related HBOV (tentatively named Macaca mulatta bocaparvovirus [MmBOV]) in fecal samples of Macaca mulatta from Guangxi Longhu Mountain, China. Additionally, we describe its complete genomic sequence and genomic characteristics. Sequence and phylogenetic analyses have shown this virus to be a novel primate BOV species.
-
Fecal samples of Macaca mulatta were collected from four distinct sites (Haijun, Lujun, Kongjun, Yingbin) in Longhu Mountain that is a local scenic spot in Guangxi Province, China. Five fecal samples were randomly collected from each site every day, when a mountain guard fed Macaca mulatta at a fixed time. In total, 400 fresh stool samples from Macaca mulatta were collected from January to May 2016. These fecal samples were transported to the Chinese Center for Disease Control and Prevention (China CDC) and stored at –80 ℃ until use. The protocol for this study was approved by the Ethics Committee of China CDC and adhered to Chinese ethics laws and regulations.
-
All 400 samples were diluted with phosphate-buffered saline (1:10 w/v), vortexed vigorously, and centrifuged at 8000 ×g for 10 min. The supernatants of every 10 samples were pooled, passed through 0.22-μm filters, and digested with DNase (Turbo DNase; Ambion, Foster City, CA, USA). Total nucleic acids were extracted in 50 μL DEPCtreated water using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The viral nucleic acid libraries were constructed by sequence-independent random reversetranscription polymerase chain reaction (RT-PCR) and then sequenced using an Illumina MiSeq 2500 platform (Illumina, San Diego, CA, USA) to yield single reads of 250 base pairs (bp) using a MiSeq Reagent Kit (v2; Illumina). Initial sequencing data were filtered and then analyzed as reported previously (Finkbeiner et al. 2008).
-
All 400 fecal samples from Macaca mulatta were screened for MmBOV by semi-nested RT-PCR using degenerate primers targeting 252 bp in the NS1 gene of MmBOV and Ex-Taq polymerase (Supplementary Table S1). The cycle conditions were as follows: 98 ℃ for 5 min, followed by 30 cycles at 98 ℃ for 20 s, 54 ℃ for 20 s, and 72 ℃ for 30 s, and final extension at 72 ℃ for 5 min. The PCR products were then visualized on 1.5% agarose gel. Sequences of positive PCR products were determined using the Big-Dye terminator cycle sequencing kit and the ABI Prism 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
-
The genomic sequence of MmBOV was amplified using the primer walking and modified RACE methods, as described previously (Kapoor et al. 2010a; Allander et al. 2005). The final acquired genomic sequence was confirmed by overlapped RT-PCR using LA-Taq DNA polymerase (Takara Bio Inc., Kusatsu, Shiga Prefecture, Japan). All primers used are listed in Supplementary Table S1.
-
DNAStar software (ver. 10) was used to compare the sequences of MmBOV with those of other BOVs. Pairwise nt and amino acid (aa) identities between MmBOV and other BOVs were calculated using DNAStar. Potential recombination was conducted on the complete genome sequences of MmBOV and those of primate BOV using Simplot software with the default setting.
-
The frequency of G+C occurrence, including GC3s, was calculated using SSE software with the default parameters. Statistics for the effective number of codons (ENC) were used to measure codon usage bias in 15 HBOVs and 4 GBOVs, as well as in the MmBOV genomes, using SSE software (Simmonds 2012).
-
Phylogenetic analyses of the almost complete MmBOV genome and the NS1, NP1, and VP1 nt sequences were conducted using the maximum likelihood method and datasets of 1, 000 replicates. The aa general time reversible substitution model and gamma distribution with invariant sites were applied using MEGA 6.0 software.
Samples
MiSeq High-Throughput Sequencing
MmBOV Screening
MmBOV Genome Sequencing
Sequence Analysis
Codon Usage Analysis
Phylogenetic Analysis
-
A total of 250 reads with a mean length of 125 bp from five pools of fecal samples identified by high-throughput sequencing had maximum nt sequence similarity to those of HBOVs, based on the analysis of customized informatics. Finally, 16 contigs (150–796 bp) were generated by assembling these positive reads, and these contigs showed ~74.8%–78.0% nt identity to HBOVs, indicating the possible presence of novel BOVs related to known HBOVs.
-
The sequence from one contig targeting the conserved NS1 gene of BOV was selected for screening for BOV primers. Seven (1.75%) of the 400 fecal samples from Macaca mulatta were positive for BOV (tentatively named MmBOV) in hemi-nested PCR. The sequences from the seven BOV-positive samples showed 100.0% nt identity with each other. The information details of positive samples are provided in a Supplementary Table S2.
-
The almost complete MmBOV nt sequence was 4, 831 nucleotides in length. Based on similarity to other known BOVs and with use of the NCBI's ORF finder, the MmBOV genome was predicted to contain three ORFs. ORF1 (638 aa) encodes the nonstructural protein NS1, ORF2 (662/533 aa) encodes the two overlapping structural proteins VP1 and VP2, and ORF3 (212 aa) encodes the highly phosphorylated nonstructural protein NP1 (Fig. 1A). The NP1 gene of MmBOV shared a short overlapping sequence with the VP1 gene, but was separated by the NS1 gene, similar to HBOVs. Also similar to HBOVs, a splicing site in the NS gene was identified in MmBOV, indicating that it also contained the spliced NS2 protein (Fig. 1B). Like HBOVs, the MmBOV genome had a characteristically low G/C content, no GC3 content, and high (> 40) ENC values, with the latter somewhat higher than those in HBOVs (Hussain et al. 2019) (Fig. 1C). Conserved motifs, including rolling circle replication, helicase, and ATPase (421–429 aa), were identified in the predicted NP1 protein of MmBOV, similar to closely related HBOVs; the Ca2+ binding loop (Y18LGPF) and the catalytic center (H41DXXY) of phospholipase A2 in the unique region of VP1 (VP1u), which are required for parvovirus infectivity (Zadori et al. 2001), were also identified in the VP1 of MmBOV (Fig. 1D). The sequence of MmBOV has been deposited in GenBank (accession numbers: MN091929).
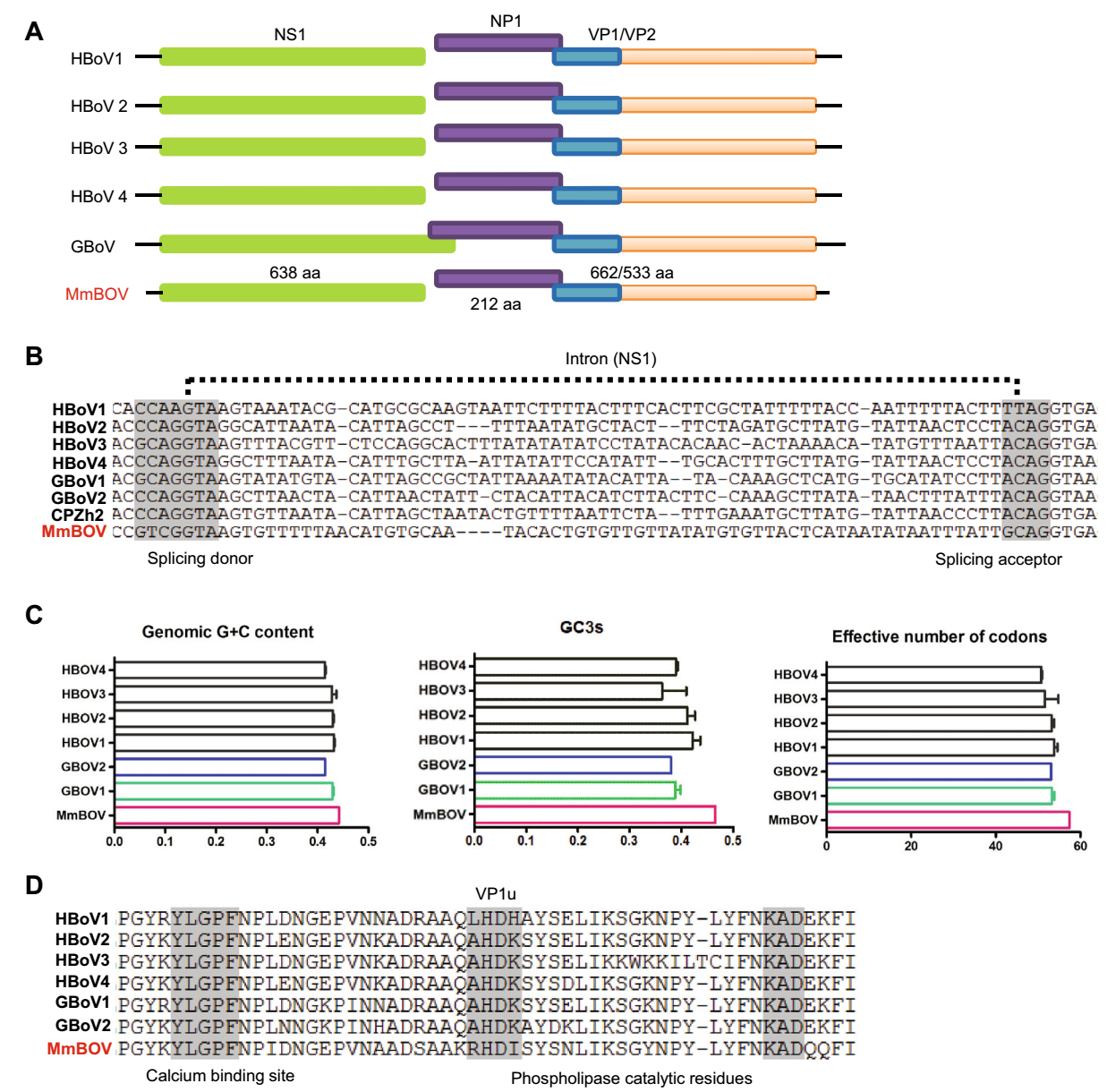
Figure 1. Genomic organization of MmBOV. (A) Genomic organization of MmBOV and known primate bocaparvoviruses. (B) Partial NS1 nucleotide sequence alignment of MmBOV and known primate bocaparvoviruses. (C) Genomic features of MmBOV and other primate bocaparvoviruses. (D) Amino acid sequence alignment of partial VP1 proteins of MmBOV and other primate bocaparvoviruses. The phospholipase A2 motif, consisting of the calcium binding region and catalytic residues, are shown. HBOV human bocaparvovirus, GBOV gorilla bocaparvovirus, CPZh2 central chimpanzee bocaparvovirus.
-
A BLASTN analysis of the complete genome revealed that MmBOV shared the highest nt identity with HBOV4 (72.0%; MG383446.1). The NS1, NP1, and VP1 sequences of MmBOV also had high degrees of similarity (68.2%/ 69.2%, 73.3%/67.6%, and 70.4%/73.1% at the nt/aa levels) with those of HBOV4 (Table 1). Table 1 lists the similarities between MmBOV and other BOVs. Simplot analysis revealed that MmBOV shared identities with human and gorilla/chimpanzee BOVs, supporting the pairwise identities shown in Table 1 and Fig. 2A. Therefore, these results suggested no obvious recombination in MmBOV, based on the known HBOV and GBOV sequences.
HBOV1 HBOV2 HBOV3 HBOV4 GBOV1 GBOV2 CPZh2 Othersa Note NS1 67.4–67.6 67.2–67.8 67.0–67.2 67.6–68.2 66.7 68.4 67.4 NT 69.7–69.8 65.8–70.8 67.5–68.3 68.3–69.2 67.7 70.5–70.6 67.0 ≤ 45.7 AA NP1 70.9–71.1 73.0–73.1 69.9–72.1 72.9–73.3 69.2 72.9 68.6 NT 63.1–63.8 66.7–66.8 62.7–67.6 66.2–67.6 61.3 67.1 60.9 ≤ 50.6 AA VP1 69.0–69.2 70.2–70.5 69.8–70.0 70.1–70.4 69.2 69.3 56.1 NT 69.7–70.0 72.1–72.5 72.3–72.7 72.8–73.1 70.5 70.3 55.2 ≤ 48.7 AA HBOV human bocaparvovirus, GBOV gorilla bocaparvovirus, CPZh2 central chimpanzee bocaparvovirus.
aAll other known bocaparvoviruses. NT and AA represent pairwise comparison of nucleotide and amino acid, respectively.Table 1. Nucleotide and amino acid identity (%) between MmBOV and other bocaparvoviruses.
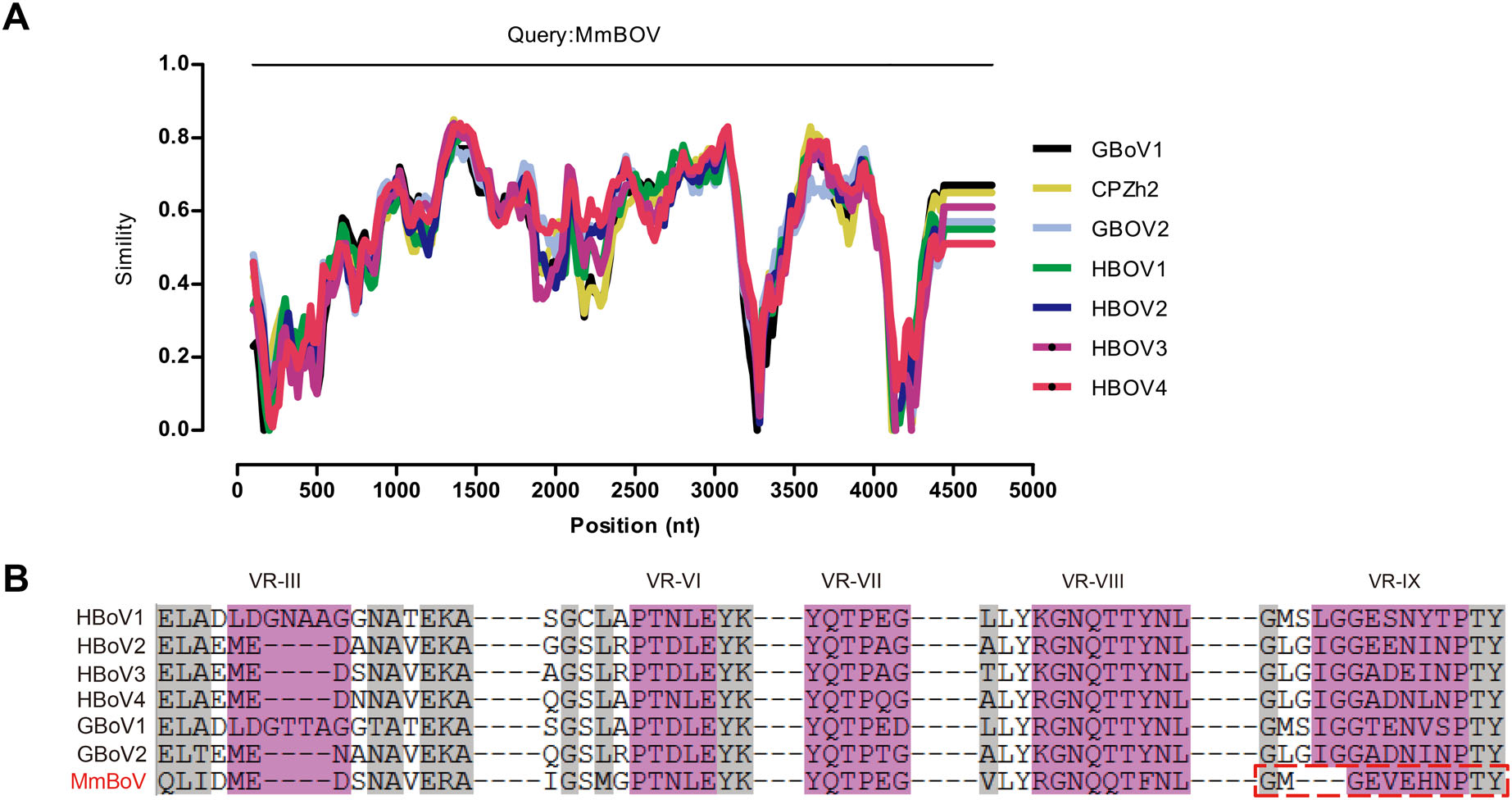
Figure 2. Comparative sequence analysis of MmBOV. A Almost complete genomic sequence identity of MmBOV and other known primate bocaparvoviruses, determined by Simplot analysis. B Variable region-based sequence alignment of MmBOV and other primate bocaparvoviruses. VR-Ⅲ, VR-Ⅵ-Ⅷ, and VR-Ⅸ are indicated in pink color. The identical residues among the human bocaparvoviruses are highlighted by gray color.
Previous structural analysis of HBOVs suggested the variable region Ⅲ (VR-Ⅲ) as a potential determinant of the tissue tropism of HBOVs, and VR-Ⅵ–Ⅷ and VR-Ⅸ as possible host range determinants (Kailasan et al. 2015; Mietzsch et al. 2017). Sequences analysis showed that the VR-Ⅲ of MmBOV was identical to that of HBOV2–4, but distinct from that of HBOV1 (Fig. 2B). In terms of host determinants, VR-Ⅵ–Ⅷ of MmBOV were similar to those of HBOVs, whereas MmBOV differed from HBOVs in the deletion of three aas in VR-Ⅸ (Fig. 2B).
-
To determine the relationship of MmBOV to other known BOVs, phylogenetic analyses were performed for the NS1, NP1, and VP1 nt sequences. From the almost complete genome, we observed that MmBOV formed a single branch between primate bocaparvovriruses and the cluster including dromedary camel/ bovine porcine BOVs in the BOV genus, but was more closely related to the Primate bocaparvovirus than to other animal BOVs (Fig. 3). Phylogenetic trees of the NS1, NP1, and VP1 genes consistently showed that MmBOV formed a monophyletic peripheral branch in the primate BOVs, but clustered most closely to those of the Primate bocaparvovirus 2 group (Fig. 4), which is consistent with our sequence analysis results.

Figure 3. Phylogenetic tree of MmBOV and other bocaparvoviruses based on almost complete nucleotide sequences. The tree of complete nucleotide sequences was reconstructed using the maximum-likelihood method with 1000 bootstrap replicates. The viruses from species Primate bocaparvovirus 1, 2, 3 are shown by green, blue and pink, respectively. The position of MmBOV is marked by ■.

Figure 4. Phylogenetic tree of MmBOV in bocaparvoviruses based on three complete coding sequences. The MmBOV trees were constructed based on the complete nucleotide sequences of NS1 (A), NP1 (B), and VP1 (C), aligned with those of other primate bocaparvoviruses using the maximum-likelihood method with datasets of 1000 replicates using MEGA 6.0 software. The viruses from species Primate bocaparvovirus 1, 2, 3 are shown by green, blue and pink, respectively. The position of MmBOV is marked by ■.
Identification of Novel BOV Sequences
Prevalence of MmBOV
Genomic Characterization of MmBOV
Sequence Analyses of MmBOV
Phylogenetic Analysis of MmBOV
-
Macaca mulatta is one of the most common primates throughout the world and thrives in protected forests, gardens, and rural villages. These animals have also been kept as domesticated pets and performing monkeys, which may increase opportunities for NHP–human virus transmission. Therefore, a thorough survey of viruses in Macaca mulatta is important to predict any threats they may pose to humans. Here, we report on the identification and the nearly complete genome sequencing of a novel BOV in the feces of wild Macaca mulatta in Guangxi, China.
In this study, the complete genome sequence of MmBOV was almost determined by walking and modified RACE methods. However, its terminal genome was not completely determined by repeated amplification, due to its terminal hairpin sequences for BOVs (Allander et al. 2005). Based on the current criteria of the ICTV (https://ictvonline.org/virusTaxonomy.asp), a novel BOV species is defined as one that shares < 85.0% aa identity in the NS1 gene with other species. Sequence analysis showed that MmBOV shared < 70.6% aa identity in the NS1 protein with other BOVs. In addition, phylogenetic analysis of the NS1, NP1, and VP1 genes showed that MmBOV formed a separate monophyletic branch in the BOV genus and was related most closely to HBOV4 in Primate bocaparvovirus 2. Furthermore, genomic analyses revealed that it displayed the HBOV characteristics of genomic organization, conserved low G/C content, and conserved weak codon usage bias (Hussain et al. 2019; Zhao et al. 2008). The MmBOV genome also had a perfectly conserved NS gene RNA splicing site, indicative of the expression of two different NS proteins, as seen in HBOVs. Based on these data, we suggest that MmBOV represents a novel species (tentatively named Primate bocaparvovirus 3). To our knowledge, all BOVs identified in NHPs to date belong to one of two genotypes in the species Primate bocaparvovirus 1/2 (Nze-Nkogue et al. 2017). This report is the first to describe a novel species of BOV in NHPs that is related to HBOVs. Its identification expands our understanding of the diversity of primate BOVs.
To date, the origins of HBOV1 and HBOV4 have not been clarified. Based on the phylogenetic analysis of the genome and the NS1, NP1, and VP1 sequences, MmBOV consistently formed a peripheral branch and clustered close to the primate BOV species. HBOV4 has been previously suggested to have diverged about 200–300 years earlier than other HBOVs (Babkin et al. 2013). In this study, MmBOV was more closely related to HBOV4 than to other BOVs from humans and NHPs, suggesting that MmBOV and HBOV4 share a common ancestor that appeared in NHPs before 200–300 years of the emergence of HBOV4. Recombination between different HBOVs plays an important role in the emergence of new genetic variants of primate BOVs (Kapoor et al. 2010b, 2011; Chong and Ng 2017; Fu et al. 2011; Song et al. 2010), but no potential recombinant was predicted for MmBOV, suggesting that MmBOV evolved from an original virus in Macaca mulatta or another NHP species. Therefore, these data strongly suggest that HBOV4 originated from primate BOVs around 200–300 years ago. Additionally, the genomic phylogenetic analysis showed MmBOV was phylogenetically between the previous primate and other mammal BOVs, which implied that MmBOV may be a special evolutionary intermediate between primate bocaparvoviruses and other mammal BOVs. Meanwhile, the relatively large distance between MmBOV and other primate BOVs also suggests that the diversity of related HBOVs in NHPs should be much greater than reported previously, and that additional novel virus species may be present in NHPs. Consequently, our findings regarding the MmBOV genome provide novel insight into the evolution of HBOVs.
MmBOV was present in ~1.75% of the fecal samples from Macaca mulatta, suggesting a relatively low prevalence. The 100% partial NS1 nt identities from the positive samples suggest that MmBOV has been circulating stably in the local Macaca mulatta population. Further studies are required to determine the diversity and epidemiology of Primate bocaparvovirus 3 in Macaca mulatta, as well as in other NHPs. HBOV infections have been linked to respiratory, enteric, and neurological diseases in humans (Manteufel and Truyen 2008; Jartti et al. 2012; Mori et al. 2013), suggesting that HBOVs cause a wide range of human illness. Gorilla Primate bocaparvovirus 1 was recently identified, and an association with gastrointestinal diseases was indicated. Comparative sequence analysis showed that MmBOV shared the VR-Ⅲ, acting as the determinant of tissue tropism, with HBOV2–4, which are related to gastrointestinal tract infections (Mietzsch et al. 2017). In this study, MmBOV was identified in stool samples from Macaca mulatta, and the symptoms that it may have caused are not known, raising the possibility that MmBOV causes gastrointestinal diseases in Macaca mulatta similar to those caused by HBOV4. In vivo studies of Macaca mulatta infection with MmBOV are required to determine the pathogenesis of this BOV.
Many related HBOVs have been found recently in NHPs, suggesting cross-species transmission of BOVs between NHPs and humans. The increasing diversity of primate BOVs in NHPs suggests that NHPs act as reservoirs for BOVs with the potential to infect humans. Moreover, sequence and phylogenetic analyses showed that MmBOV is related most closely to HBOV2/4. Although sequence analysis showed that MmBOV was somewhat distinct in terms of VR-Ⅸ as the possible host range determinant (Mietzsch et al. 2017), the mutation accumulation rates of BOVs are high and similar to the evolutionary rate of RNA viruses (Shackelton et al. 2005; Hoelzer et al. 2008), which suggests that MmBOV has the capacity to adaptively infect new host species. Therefore, the possibility of cross-species transmission of MmBOV from NHPs to humans cannot be discounted. With the expansion of human activity, the likelihood of human– NHP interaction has increased, creating a route for the rapid spread of viral infections, such as human Ebola/ Marburg viruses and the human immunodeficiency virus, from NHPs to humans (Gonzalez et al. 2007; Keele et al. 2006). Therefore, studies of the potential ability of related MmBOVs to infect humans are necessary, including serological epidemiological assays on humans living in proximity to NHP populations to test for infection risk. Furthermore, a survey of BOV infections in wild NHPs is needed to improve our understanding of the evolution and safe management of wild NHPs.
-
This study was funded by the National Science and Technology Major Project (2018ZX10301408-001).
-
ZD designed and guided the study. YA performed the experiments. YA contributed in the collection and analysis of the data. YA wrote the manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
The study was approved by the Chinese CDC's Ethics Committee on the use of animals and complied with Chinese ethics laws and regulations.
















 DownLoad:
DownLoad: