HTML
-
Prototype foamy virus (PFV), first known as human foamy virus (HFV), belongs to Spumaretovirinae from the Retroviridae family (Khan et al. 2018). PFV was first discovered in the 1970s and was isolated from a nasopharyngeal carcinoma patient in Kenya (Achong et al. 1971). Although PFV causes a lifelong infection, no clinical symptoms have been reported in animals or humans infected with the virus (Effantin et al. 2016; Xu et al. 2019). Compared with the genomes of other retroviruses, PFV has the longest genome, which may be able to carry large exogenous genes. In addition, PFV has shown potential therapeutic results in preclinical studies using animal model systems (Armbruster et al. 2014; Counsell et al. 2018).
PFV contains a unique internal promoter (IP), which makes its replication and expression complicated (Lochelt et al. 1993). IP plays a crucial role in initiating transcription during the early period of PFV infection (Ma et al.2014). IP activation triggers the expression of the viral transcriptional transactivator (Tas), which is essential for viral replication and gene expression. Elevated Tas results in establishing a positive feedback loop and upregulates IP transcriptional activity (Campbell et al. 1994; Hu et al. 2014; Ma et al. 2014). The Tas protein transactivates the long terminal repeat (LTR) promoter, which is required for viral replication (Hu et al. 2014; Goodman et al. 2018). In recent years, studies of Tas have focused on binding proteins and on their effects on viral replication. Little is known about the regulation of Tas expression by the basal transcription of PFV IP.
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides without a protein-coding ability (Guttman et al. 2009; Mattick and Rinn 2015). LncRNAs are confirmed to be involved in the processes of various viral infections (Shi et al. 2016; Wang et al. 2017; Li et al. 2018; Ji et al. 2019). Recently, we revealed the differential lncRNA expression profiles in PFV-infected cells (Xu et al. 2017) and found that the expression of lncRNA lnc-RP5-1086D14.3.1-1:1 (lncRP5) was downregulated in PFV-infected cells, which indicates that lncRNA lnc-RP5 might play an important role during PFV infection. In this study, we report that lnc-RP5 promotes the expression of PFV Tas through the miR-129-5p/Notch1/PFV IP axis.
-
293T cells (human embryonic kidney cells) were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) (HyClone, Utah, USA). HT1080 cells (human fibrosarcoma cells) were cultured in minimum essential medium (MEM) supplemented with 10% FBS (HyClone, Utah, USA). Both cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). PFV was prepared by transfecting 293T cells with the provirus plasmid pHSRV13 (German Cancer Research Center, Heidelberg, Germany) (Lochelt et al. 1991) by the modified calcium phosphate method (Lochelt et al. 1991; Yuan et al. 2017). The transfected cells and the culture medium were frozen and thawed for three cycles to release the virus after 48 h. To prepare cell-free virus stocks, the culture supernatants were centrifuged at 4000 9 g for 10 min, filtered through a 0.22 lm pore size filter membrane and stored at -80 ℃. PFV titers were determined by infecting PFV indicator cells (Tai et al. 2001; Dong et al. 2015).
-
To generate the plasmid encoding lnc-RP5, we first cloned a full-length fragment of the lnc-RP5 transcript from the 293T cell cDNA library. The primers used for plasmid construction are listed in Supplementary Table S1. Then, a full-length fragment of lnc-RP5 was inserted into the pcDNA3.1(-) vector using the Xho I and Kpn I restriction sites. The restriction enzymes were purchased from Takara Bio (Dalian, China), and the empty vector pcDNA3.1(-) plasmids were purchased from Addgene (Massachusetts, USA).
-
The total RNA was extracted from cells with TRIzol Reagent (Invitrogen, California, USA), and the RNA was reverse transcribed into cDNA using a Reverse Transcription Kit (Thermo, Massachusetts, USA) according to manufacturer's protocols. qRT-PCR experiments were performed using a SYBR green mix (Toyobo, Osaka, Japan), and the primers used in the study are listed in Supplementary Table S1. The conditions for measuring the miRNA expression levels were 95 ℃ for 20 s and 40 cycles of 95 ℃ for 10 s, 60 ℃ for 20 s and 70 ℃ for 10 s. To determine the Notch1, Gag and Tas expression levels, the conditions were 95 ℃ for 30 s and 40 cycles of 95 ℃ for 10 s, 60 ℃/58 ℃/55 ℃ for 15 s and 72 ℃ for 20 s. Actin was used as internal control for Notch1, Gag and Tas. U6 snRNA was used as internal control for miR-129-5p.
-
293T cells or HT1080 cells were seeded into 6- or 12-well plates. The cells were transfected with plasmids, siRNA or miRNA using Lipofectamine 2000 (Invitrogen, USA). The following plasmids were used in this study: IP-Luc, RLTK, TK-Tas, lnc-RP5-pcDNA3.1(-), pcDNA3.1(-), premiR129-pHage, pHage and Notch1-pcDNA3.1(-). The lncRP5 siRNAs (siRNA-209, siRNA-1044, siRNA- 1857), Notch1 siRNAs (siRNA-780, siRNA-2010, and siRNA- 3916) and negative control siRNA were purchased from Gene Pharma (Shanghai, China), and the primers for the siRNAs used in the study are listed in Supplementary Table S2. The miRNA-129-5p inhibitor, the negative control for the inhibitor and the primers for miRNA-129-5p were purchased from Ribo Biotech (Guangzhou, China). All of them were used according to the manufacturer's instructions. Twenty-four hours after transfection, the cells were infected with PFV. After 1.5 h of infection, the supernatant from the infected cells was replaced with culture medium and was maintained for 24–48 h.
-
To determine whether the candidate genes influence PFV IP activity, 50 ng IP-Luc, 3 ng RL-TK, 60 ng TK-Tas, 630 ng candidate gene and 630 ng negative control for the candidate gene were transfected into 293T cells in 24-well plates. Each experimental group contained sextuplicate or octuplicate samples. The Dual-Glo Luciferase Assay System (Promega, USA) was used to test the PFV IP activity according to the firefly and Renilla luciferase activities.
-
293T cells or HT1080 cells were washed with PBS twice or thrice. The cells were lysed with RIPA buffer containing phenylmethanesulfonyl fluoride (PMSF, 100:1) on ice for 15–20 min. The cell lysates were then mixed with SDS loading buffer. After boiling the samples for 10 min, the samples were separated by 10% or 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, MMAS, USA). The membrane was blocked in 5% non-fat milk for 2 h at room temperature. Then, the membrane was incubated with a primary antibody overnight at 4 ℃. After washing with TBST, a secondary antibody was applied to the membrane, and the membrane was incubated for 2 h at room temperature. Finally, enhanced chemiluminescence reagents (ECL, Thermo Scientific, Waltham, MA, USA) were used to detect the proteins. The primary antibodies used were antiNotch1 (Cell Signaling Technology, Massachusetts, USA), anti-Actin (Abcam, Cambridge, UK), anti-Gag and antiTas antibodies. The anti-Tas and anti-Gag antibodies were prepared by immunizing rabbits with the Tas protein and Gag protein. The secondary antibody was goat anti-rabbit (Proteintech Group, Chicago, USA).
-
All reactions were independently repeated three times. All data were analyzed with Prism 5 software and Student's t test. The results were represented by P values, and P < 0.05 was considered to be statistically significant.
Cell and Virus Cultures
Plasmid Construction
RNA Extraction and qRT-PCR
Plasmids, siRNA, miRNA Transfection and Virus Infection
Luciferase Assays
Western Blot Analysis
Statistical Analysis
-
Lnc-RP5 was predicted to be an intergenic lncRNA with a total length of 1988 bp and three exons (LNCipedia website, http://www.lncipedia.org/). Because lnc-RP5 is significantly downregulated during PFV infections (Xu et al. 2017) and PFV IP plays a crucial role during PFV early transcription (Linial 1999; Yang et al. 2000), we wondered whether lncRP5 had an effect on PFV replication. First, we cloned the lnc-RP5 cDNA and reconstructed the overexpression plasmid lnc-RP5-pcDNA3.1(-). After transfection with lnc-RP5- pcDNA3.1(-), the expression of lnc-RP5 in 293T cells was enhanced more than 70, 000-fold compared with that of the control group (Fig. 1A). To assess the effect of lnc-RP5 on PFV IP, the plasmids PFV IP-Luc, RL-TK, TK-Tas (Dong et al. 2015) and lnc-RP5-pcDNA3.1(-) were transfected into 293T cells. We found that lnc-RP5 overexpression enhanced the activity of PFV IP by approximately 2.5-fold compared with that of the control group in luciferase assays (Fig. 1B). These results suggest that lnc-RP5 effectively enhances the activity of PFV IP.
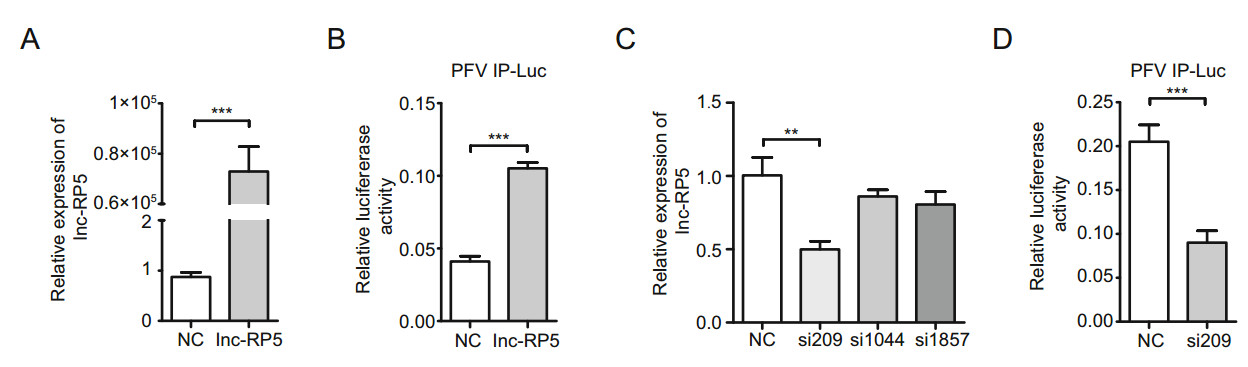
Figure 1. Lnc-RP5 enhances the activity of PFV IP. A 293T cells were seeded in 6-well plates, and then transfected with the 4 lg plasmid lnc-RP5-pcDNA3.1(-) when cells were at approximately 60% confluence. Forty-eight hours after transfection, the mRNA levels of lnc-RP5 were determined by qRT-PCR assays. B The overexpression of lnc-RP5 enhanced the activity of PFV IP, as detected by luciferase assays. 293T cells were transfected with the 630 ng plasmid lnc-RP5-pcDNA3.1(-) together with 50 ng PFV IP-Luc, 3 ng RL-TK, and 60 ng TK-Tas when cells were seeded at approximately 60% confluence in 24-well plates. Then, the activities of PFV IP were tested by luciferase assays. C The mRNA expression of lnc-RP5 was significantly decreased in the cells treated with siRNA-209. The 5 μL siRNA-209, siRNA-1044, siRNA-1857 and the negative control siRNA were respectively transfected into 293T cells when cells were seeded at approximately 60% confluence in 12-well plates. The mRNA levels of lnc-RP5 were determined by qRT-PCR assays. D siRNA-209 suppressed the activity of PFV IP. The 630 ng siRNA- 209 was cotransfected with 50 ng PFV IP-Luc, 3 ng RL-TK, and 60 ng TK-Tas into 293T cells when cells were seeded at approximately 60% confluence in 24-well plates. All data were analyzed with Prism 5 software and Student's t test and are the mean values ± SDs; **P < 0.01, ***P < 0.001.
To further determine the role of lnc-RP5 in PFV replication, we constructed three lnc-RP5 siRNAs (siRNA-209, siRNA-1044, and siRNA- 1857) to downregulate lnc-RP5 expression. The most effective silencer for lnc-RP5 was siRNA-209 because siRNA-209 treatment resulted in the lowest level of lnc-RP5 expression (an approximately 50% reduction in lnc-RP5 expression, Fig. 1C). Therefore, siRNA-209 was selected and used in the following experiments. As shown in Fig. 1D, the transcriptional activity of PFV IP was inhibited in lnc-RP5-silenced cells, which further confirmed that lnc-RP5 enhances PFV IP activity.
-
Next, we assessed the effects of lnc-RP5 on the expression of PFV Tas. We used HT1080 cells in a PFV in vitro infection model because these HT1080 cells are susceptible to PFV and have been used to investigate PFV gene expression (Hamann et al. 2014b; Effantin et al. 2016). By qRT-PCR assays, overexpressed lnc-RP5 enhanced the mRNA expression of Tas by approximately 2.3-fold compared with that of the control group (Fig. 2A). By Western blot assays, overexpressed lnc-RP5 promoted the expression of Tas by approximately 1.7-fold compared with that of the control group at the protein level (Fig. 2B). These results demonstrate that lnc-RP5 could increase the expression of Tas. Consistent with the results of Fig. 2A and 2B, the mRNA level of Tas were decreased by approximately 53% in lnc-RP5-silenced group compared with that of the control group, as detected by qRT-PCR assays (Fig. 2C). The protein level of Tas was decreased by 50% in the siRNA-209 treatment group compared with that of the control group in the Western blot assay (Fig. 2D). These results indicate that lnc-RP5 might promote PFV Tas expression.
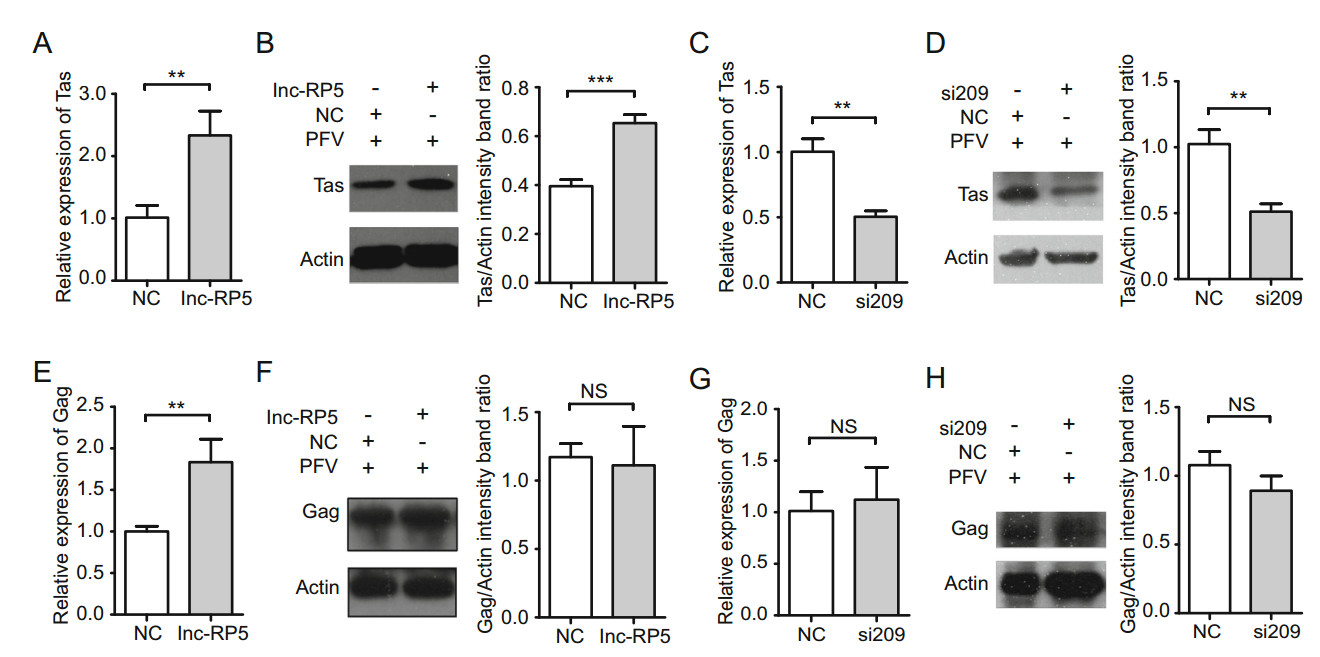
Figure 2. Lnc-RP5 promotes the expression of PFV Tas but has no effect on the expression of PFV Gag. The 4 lg overexpressed lnc-RP5 plasmid and 10 μL siRNA-209 were respectively transfected into HT1080 cells for 24 h when cells were seeded at approximately 60% confluence in 6-well plates. Then, the cells were infected with PFV for an additional 1.5 h. After washing, the cells were cultured for 48 h. RNA and protein were extracted for the detection of Tas/Gag mRNA and protein. A The mRNA level of PFV Tas was increased in lnc-RP5-overexpressing and PFV-infected cells in a qRT-PCR assay. B The protein level of PFV Tas was increased in lnc-RP5- overexpressing and PFV-infected cells in a Western blot assay. C The mRNA level of PFV Tas was decreased in lnc-RP5-silenced and PFV-infected cells in a qRT-PCR assay. D The protein level of PFV Tas was decreased in lnc-RP5-silenced and PFV-infected cells in a Western blot assay. E The mRNA level of PFV Gag was increased in lnc-RP5-overexpressing and PFV-infected cells in a qRT-PCR assay. F The protein level of PFV Gag has no significant difference in lnc-RP5-overexpressing and PFV-infected cells in a Western blot assay. G The mRNA level of PFV Gag has no significant difference in lnc-RP5-silenced and PFV-infected cells in a qRT-PCR assay. H The protein level of PFV Gag has no significant difference in lnc-RP5- silenced and PFV-infected cells in a Western blot assay. Left panel, the presentative blots. Right panel, pooled data. All reactions were independently repeated three times. All data were analyzed with Prism 5 software and Student's t test as the mean values ± SDs; **P < 0.01, ***P < 0.001; NS, no significant difference.
PFV Gag was reported to play an important role in many stages of the PFV replication cycle. PFV Gag regulates viral assembly, interacts with host proteins, modulates expressions of viral genes, and participates in viral trafficking and budding (Reh et al. 2013; Hamann et al. 2014a; Lesbats and Parissi 2018). So we think that a late event of the viral cycle could be reflected by the effect of lnc-RP5 on Gag replication. As shown in Fig. 2E, overexpressed lnc-RP5 enhanced the mRNA expression of PFV Gag by approximately 1.8-fold compared with that of the control group by qRT-PCR assays. However, overexpressed lncRP5 could not promote the protein expression of PFV Gag compared with that of the control group by Western blot assays (Fig. 2F). At the same time, lnc-RP5 si209 has no effect on the expression of PFV Gag on mRNA level and protein level compared with that of the control group in qRT-PCR assays and Western blot assays (Fig. 2G, 2H). These results demonstrate that lnc-RP5 has no effect on the expression of PFV Gag. Together with our current results, lnc-RP5 plays an important role in early stage rather than late stage of the PFV replication cycle.
-
LncRNA may act as a competing endogenous RNA (ceRNA) to influence miRNA expression (Zhen et al. 2016; Chen et al. 2018; Yu et al. 2019). To look for potential miRNAs involved in lnc-RP5 expression, we predicted the miRNAs that hybridize with lnc-RP5 by using the bioinformatics software RNA22 v2 (https://cm.jefferson.edu/rna22/Interactive/). The predicted results indicate that lnc-RP5 might bind to miR-129-5p at miRNA positions 144-164, 1341-1361, 1348-1368 or 1768-1789 (Fig. 3).
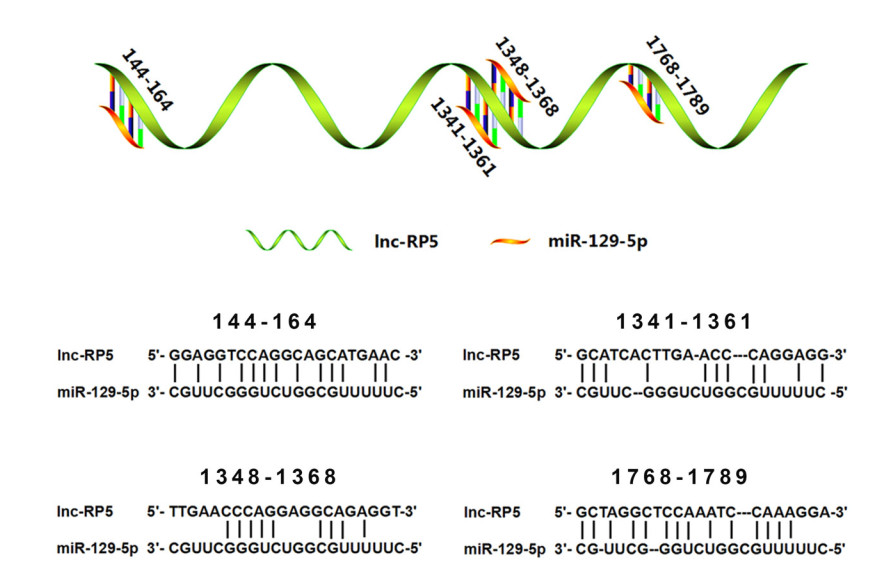
Figure 3. The binding sites of miR-129-5p to lnc-RP5 were predicted. The predicted results showed that lnc-RP5 may base pair with miR-129-5p at the miRNA positions 144–164, 1341–1361, 1348–1368 or 1768–1789.
To measure the effects of lnc-RP5 on miR-129-5p expression, the plasmid lnc-RP5-pcDNA3.1(-) was transfected into 293T cells, and the expression of miR-129-5p was tested by qRT-PCR assays. The results showed that overexpressed lnc-RP5 promoted the expression of miR- 129-5p by approximately 2.5-fold compared with the expression in the control group (Fig. 4A). Consistent with the results from lnc-RP5 overexpression, miR-129-5p expression was decreased by approximately 50% in the lnc-RP5-silenced group compared with that in the control group (Fig. 4B). These results demonstrate that miR-129-5p expression is promoted by lnc-RP5.
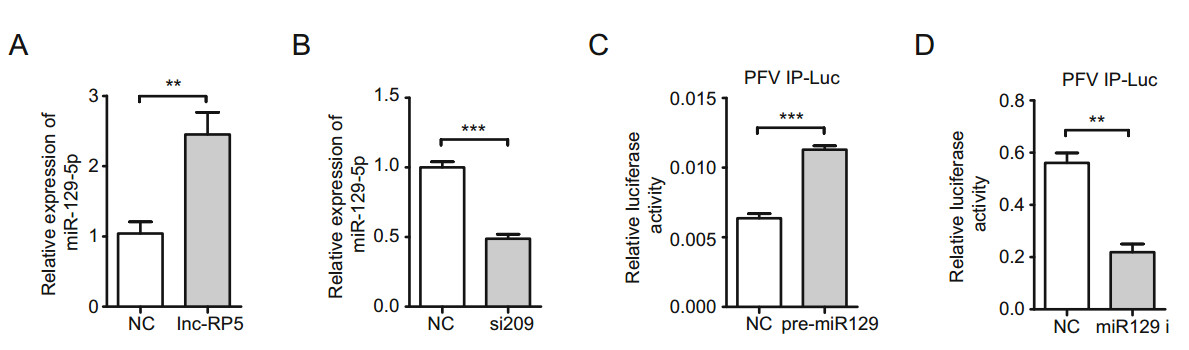
Figure 4. miR-129-5p is involved in the lnc-RP5-mediated promotion of PFV IP activity. A The overexpression of lnc-RP5 promoted miR- 129-5p expression. The 4 lg plasmid lnc-RP5-pcDNA3.1(-) was transfected into 293T cells when cells were seeded at approximately 60% confluence in 6-well plates. Forty-eight hours after transfection, the expression of miR-129-5p was determined by qRT-PCR assays. B Lnc-RP5 siRNA-209 decreased the expression of miR-129-5p. The 10 μL siRNA-209 was transfected into 293T cells when cells were seeded at approximately 60% confluence in 6-well plates. Forty-eight hours after transfection, the expression of miR-129-5p was determined by qRT-PCR assays. C The overexpression of miR-129-5p enhanced the activity of PFV IP. The 630 ng miR-129-5p precursor plasmids (encoding miR129) were transfected into 293T cells together with 50 ng plasmids PFV IP-Luc, 3 ng RL-TK, and 60 ng TK-Tas when cells were seeded at approximately 60% confluence in 24-well plates. Forty-eight hours after transfection, the transcriptional activity of PFV IP was measured by the luciferase reporter assay. D The miR-129-5p inhibitor decreased the activity of PFV IP. The 630 ng miR-129 i was transfected into 293T cells together with 50 ng plasmids PFV IP-Luc, 3 ng RL-TK, and 60 ng TK-Tas when cells were seeded at approximately 60% confluence in 24-well plates. Forty-eight hours after transfection, the transcriptional activity of PFV IP was measured by the luciferase reporter assay. All reactions were independently repeated three times. All data were analyzed with Prism 5 software and Student's t test as the mean values ± SDs; **P < 0.01, ***P < 0.001.
Furthermore, we explored the relationship between miR- 129-5p and PFV IP. The miR-129-5p precursor plasmids (encoding miR129) or the miR-129-5p inhibitor (miR-129 i) were transfected into 293T cells together with the PFV IP-Luc, RL-TK, and TK-Tas plasmids, and the transcriptional activity of PFV IP was determined by luciferase assays (Dong et al. 2015). The results indicated that the transfection of the miR-129-5p precursor plasmids caused a 1.8-fold increase in PFV IP activity compared with that in the control group (Fig. 4C), while miR-129 i decreased the PFV IP activity by approximately 60% (Fig. 4D). Together, these results suggest that miR-129-5p is involved in the lnc-RP5- mediated promotion of PFV IP activity.
-
Because the lnc-RP5/miR-129-5p/PFV IP axis may promote the expression of PFV Tas (Figs. 1, 2, 3, 4), we asked whether some host proteins were involved in the axis. In our previous study, miR-129-5p was identified as a regulator that suppressed Notch1 protein expression (Chen et al. 2016). Notch1 protein has also been confirmed to be involved in virus replication or tumorigenesis in virusinfected cells (Kongkavitoon et al. 2016; Su et al. 2018; Peng et al. 2019); therefore, we assessed the effects of lncRP5 on Notch1 expression. The results from qRT-PCR assays and Western blot assays showed that both the mRNA and protein levels of Notch1 were significantly decreased by approximately 40% in lnc-RP5-overexpressing groups compared with those in the control groups (Fig. 5A, 5B). To further confirm the lnc-RP5-mediated regulation of Notch1, siRNA-209 was transfected into 293T cells. As expected, both the mRNA and protein levels of Notch1 were increased by approximately 2-fold in lncRP5-silenced groups compared with the levels in the control groups (Fig. 5C, 5D). These results demonstrate that lnc-RP5 negatively regulates Notch1 expression.
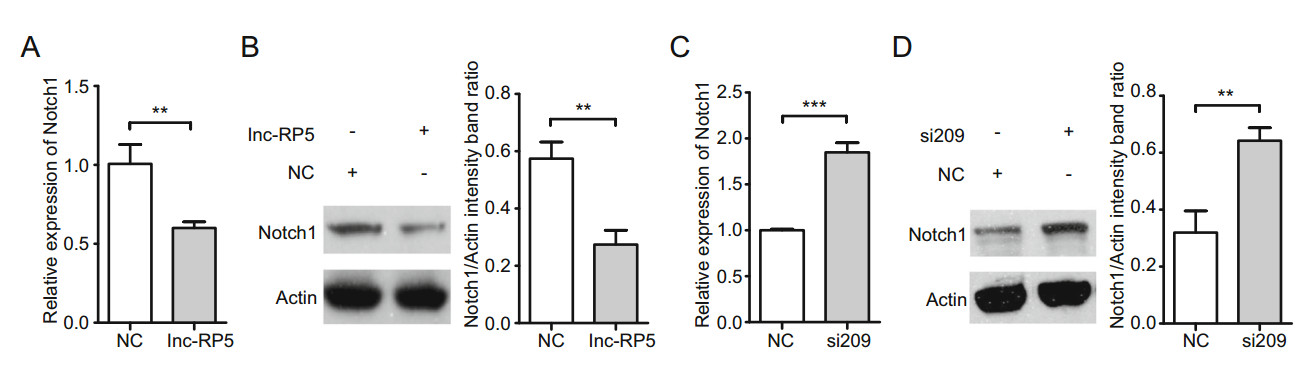
Figure 5. Lnc-RP5 suppresses Notch1 expression. A, B The 4 lg plasmid lnc-RP5-pcDNA3.1(-) was transfected into 293T cells when cells were seeded at approximately 60% confluence in 6-well plates. Forty-eight hours after transfection, the mRNA level and protein level of Notch1 were determined by qRT-PCR assays and Western blot assays. A The mRNA level of Notch1. B The protein level of Notch1. Left panel, the presentative blots. Right panel, pooled data. C, D The 10 μL lnc-RP5 siRNA-209 was transfected into 293T cells when cells were seeded at approximately 60% confluence in 6-well plates. Fortyeight hours after transfection, the mRNA level and protein level of Notch1 were determined by qRT-PCR assays and Western blot assays. C The mRNA level of Notch1. D The protein level of Notch1. Left panel, the presentative blots. Right panel, pooled data. All reactions were independently repeated three times. All data were analyzed with Prism 5 software and Student's t test as the mean values ± SDs; **P < 0.01, ***P < 0.001.
-
To investigate the effects of Notch1 on PFV IP, the plasmid encoding Notch1 was transfected into 293T cells together with plasmids PFV IP-Luc, RL-TK and TK-Tas, and the transcriptional activity of PFV IP was determined by luciferase assays. As shown in Fig. 6, the activity of PFV IP was decreased by approximately 60% in the Notch1- overexpressing group compared with that in the control group (Fig. 6A). To assess the effects of Notch1 on PFV Tas, Notch1-overexpressing HT1080 cells were infected with PFV. The mRNA and protein levels of PFV Tas were determined (Fig. 6B, 6C). The mRNA expression of PFV Tas was inhibited by the overexpression of Notch1 in the group and decreased by approximately 43% compared with that in the control group (Fig. 6B). Consistent with the mRNA level of Tas, the overexpression of Notch1 downregulated the expression of PFV Tas protein (approximately a 30% decrease) compared with that in the control group (Fig. 6C). These results indicate that the overexpression of Notch1 suppresses the activity of PFV IP and the expression of PFV Tas.
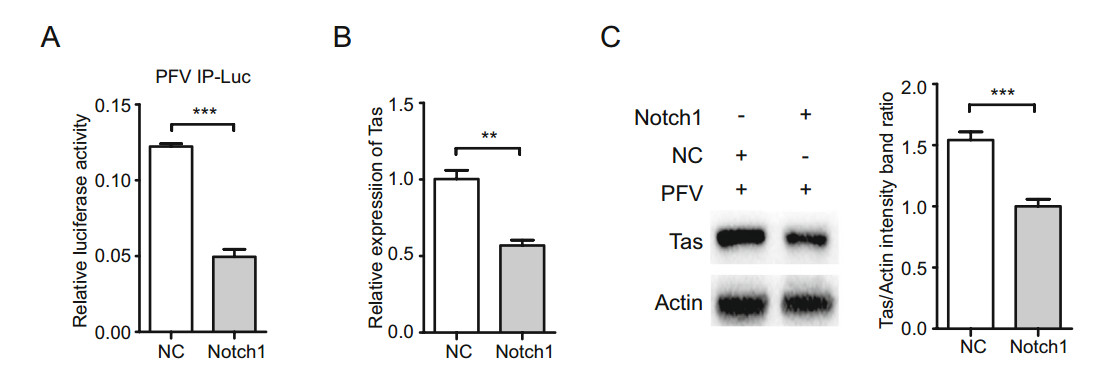
Figure 6. Notch1 suppresses the activity of PFV IP and the expression of PFV Tas. A The activity of PFV IP was suppressed in Notch1- overexpressing cells. The 630 ng plasmid (Notch1-pHage) encoding Notch1 was transfected with 50 ng PFV IP-Luc, 3 ng RL-TK, and 60 ng TK-Tas into 293T cells when cells were seeded at approximately 60% confluence in 24-well plates. Then, the activity of PFV IP was measured by luciferase assays. B, C The 4 lg plasmid Notch1- pHage was transfected into HT1080 cells when cells were seeded at approximately 60% confluence in 6-well plates. Twenty-four hours after transfection, HT1080 cells were infected with PFV. Forty-eight hours after infection, the mRNA level and protein level of PFV Tas were determined by qRT-PCR assays and Western blot assays. B The mRNA level of PFV Tas. C The protein level of PFV Tas. Left panel, the presentative blots. Right panel, pooled data. All reactions were independently repeated three times. All data were analyzed with Prism 5 software and Student's t test as the mean values ± SDs; **P < 0.01, ***P < 0.001.
To further determine the role of Notch1 in PFV replication, we constructed three Notch1 siRNAs (siRNA-780, siRNA-2010, and siRNA-3916) to downregulate Notch1 expression. Notch1 siRNA-780, siRNA-2010, and siRNA- 3916 respectively downregulated the mRNA expression of Notch1 by approximately 40%, 57% and 49% compared with that of the control group by qRT-PCR assays (Fig. 7A). The most effective silencer for Notch1 was siRNA-2010, therefore, siRNA-2010 was selected and used in the following experiments. As shown in Fig. 7B, the activity of PFV IP was increased by approximately 2.7-fold in the Notch1-silencing group compared with that in the control group. To assess the effects of silencing Notch1 on PFV Tas, Notch1-silencing HT1080 cells were infected with PFV. The mRNA and protein levels of PFV Tas were determined (Fig. 7C, 7D). The mRNA expression of PFV Tas was enhanced by Notch1 siRNA-2010 approximately 2.3-fold compared with that in the control group (Fig. 7C). Consistent with the mRNA level of Tas, Notch1 siRNA- 2010 promoted the expression of PFV Tas protein approximately 2.3-fold compared with that in the control group (Fig. 7D). These results indicate that silencing Notch1 promoted the activity of PFV IP and the expression of PFV Tas. Together, these results suggest that Notch1 suppresses the activity of PFV IP and the expression of PFV Tas.
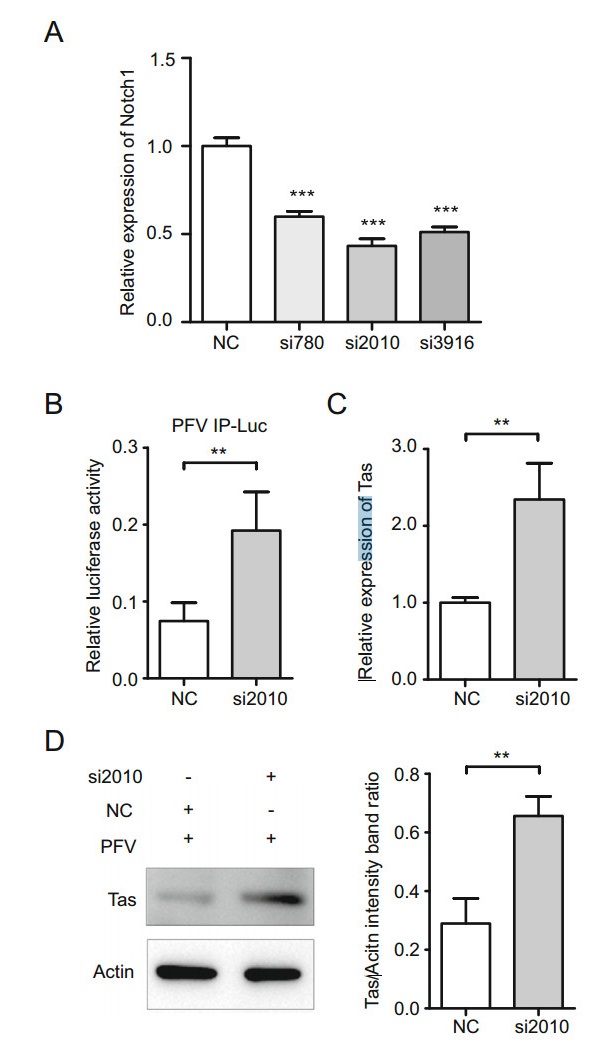
Figure 7. Silencing Notch1 promotes the activity of PFV IP and the expression of PFV Tas. A The most effective silencer for Notch1 was siRNA-2010. The 2.5 μL siRNA-780, siRNA-2010, siRNA-3916 and the negative control siRNA were respectively transfected into 293T cells when cells were seeded at approximately 60% confluence in 12-well plates. The mRNA levels of Notch1 were determined by qRT-PCR assays. B The activity of PFV IP was enhanced in Notch1- silencing cells. The 630 ng Notch1 siRNA-2010 was transfected with 50 ng PFV IP-Luc, 3 ng RL-TK, and 60 ng TK-Tas into 293T cells when cells were seeded at approximately 60% confluence in 24-well plates. Then, the activity of PFV IP was measured by luciferase assays. C, D The 5 μL Notch1 siRNA-2010 was transfected into HT1080 cells when cells were seeded at approximately 60% confluence in 6-well plates. Twenty-four hours after transfection, HT1080 cells were infected with PFV. Forty-eight hours after infection, the mRNA level and protein level of PFV Tas were determined by qRT-PCR assays and Western blot assays. C The mRNA level of PFV Tas. D The protein level of PFV Tas. Left panel, the presentative blots. Right panel, pooled data. All reactions were independently repeated three times. All data were analyzed with Prism 5 software and Student's t test as the mean values ± SDs; ** P < 0.01, *** P < 0.001.
Lnc-RP5 Enhances the Transcriptional Activity of PFV IP
The Expression of PFV Tas Is Promoted by lnc-RP5
miR-129-5p Is Involved in the lnc-RP5-mediated Promotion of PFV IP activity
Lnc-RP5 Downregulates Notch1 Expression
Notch1 Suppresses the Activity of PFV IP and the Expression of PFV Tas
-
LncRNAs have functions that include transcription modulation, signal transduction, molecular scaffolds, nuclear organization, and epigenetic regulation (Clark and Mattick 2011; Wang et al. 2016; Yan et al. 2018; Wu et al. 2019). It has been reported that the lncRNA NRON induces the degradation of the viral protein Tat to repress HIV-1 replication during viral infection (Li et al. 2016). In addition, lncRNA GAS5 could inhibit HCV replication by binding viral NS3 protein (Qian et al. 2016). However, studies on lncRNA roles during viral infection are still limited. In this study, we reported that lnc-RP5 binds to miR-129-5p to enhance the activity of PFV IP and the expression of Tas (Figs. 1, 2, 4), while the host protein Notch1 hinders the lnc-RP5/miR-129-5p-mediated promotion of PFV IP and Tas (Figs. 5, 6, 7). To our knowledge, this is the first report about lnc-RP5 function. Whether lnc-RP5 has effects on the expression of other PFV genes and whether lnc-RP5 regulates host gene transcription should be investigated in the future.
It was reported that PFV Tas plays important roles in initiating the transcription of PFV proteins (Campbell et al. 1994; Mergia 1994; Hu et al. 2014) and has functions in viral persistence or latency (Linial 2000). The transcription of viral Tas was initiated by PFV IP in the early period of virus infection. Tas could use feedback to activate IP to increase its synthesis level (Campbell et al. 1994). When the protein level of Tas accumulates to a certain extent, it transactivates foamy virus LTR to express viral structural proteins (Mergia 1994). Most studies on Tas focus on interfering with its function to suppress the expression of viral structural proteins (Tan et al. 2008; Hu et al. 2014), but the regulation of Tas expression remains elusive. Previously, our group reported that Tas expression was downregulated by a cellular protein, E3 ubiquitin ligase Pirh2 (Dong et al. 2015). In the current study, we reveal that the novel noncoding RNA lnc-RP5 binds to miR-129- 5p to regulate the expression of PFV Tas through the miR- 129-5p/Notch1/PFV IP axis, which suggests a complicated mechanism and the regulation of PFV transcription during the early period of viral infection.
It has been reported that miR-129-5p could downregulate human papillomavirus E6 and E7 viral gene expression (Zhang et al. 2013). Zhang et al. showed that the direct downstream target of miR-129-5p is the SP1 transcription factor. SP1 is a sequence-specific DNAbinding protein that contains a miR-129-5p binding site in its 30 UTR. In our current study, we report that miR-129- 5p is involved in the promotion activity of PFV IP, indicating that miR-129-5p might widely participate in the transcriptional regulation of various viral genes. Whether SP1 is involved in the promotion of PFV IP activity or the expression of Tas needs to be investigated in future studies.
An aberration in the Notch signaling pathways has been reported to be linked to various virus-related tumors (Kongkavitoon et al. 2016; Su et al. 2018; Peng et al. 2019). For example, HBV HBx could activate Notch signaling via the DII4/Notch1 signaling pathway (Kongkavitoon et al. 2016). The activation or loss of Notch1 influences HPV-induced oral tumorigenesis (Zhong et al. 2015). Notch1 protein has also been shown to activate EBV latent promoters in different cell lines (Cotter et al. 2000). Our results demonstrate that Notch1 downregulates PFV IP and Tas expression, indicating that the Notch1 protein might suppress PFV replication and contribute to asymptomatic viral infections.
Together, in this study, we demonstrate a novel mechanism of lnc-RP5 in the regulation of the expression of PFV Tas by the miR-129-5p/Notch1/PFV IP axis (Fig. 8). Lnc-RP5 binds to miR-129-5p, which enhances the activity of PFV IP and the expression of Tas. The host protein Notch1 acts as a negative regulator, and is involved in downregulating the activity of PFV IP and the expression of Tas. And Notch1 is repressed by lnc-RP5 and miR-129- 5p.
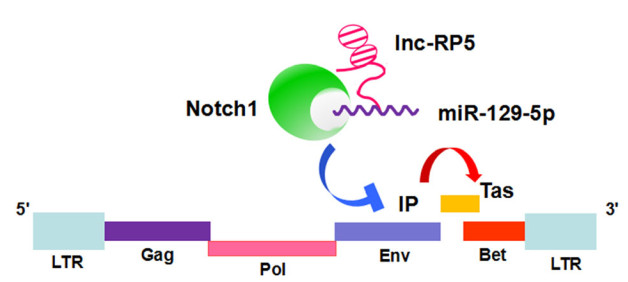
Figure 8. Lnc-RP5 could enhance the expression of miR-129-5p. Then, miR-129-5p was demonstrated to suppress the expression of Notch1. Furthermore, Notch1 represses the activity of PFV IP and the expression of Tas. Consistent with these results, lnc-RP5 has been demonstrated to promote the activity of PFV IP and the expression of Tas. Taken together, lnc-RP5 could promote the expression of the PFV transactivator Tas by regulating the miR-129-5p/Notch1/PFV IP axis.
-
This work was supported by the National Natural Sciences Foundation of China (Nos. 81641093, 81371790, 81371422, 81571481 and 81701571), the Fundamental Research Funds for the Central Universities of China and the Translational Medical Research Fund of Wuhan University School of Medicine.
-
WL and SX designed the experiments, analyzed the data and wrote the paper. SX, LC and YT carried out the experiments. PY, JY, YZ, LH, ZL and YS offered some experimental materials. SH, JY, QP, BP, XH and WL supervised this study and checked and finalized the manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: