HTML
-
Rotavirus (RV) is the largest cause of acute gastroenteritis in infants and young children, and exceeds the calicivirus including norovirus and sapovirus (Nguyen et al. 2007; Patel et al. 2008; Qiao et al. 1999; Tran et al. 2010). RV infection caused 128, 500 deaths among children younger than 5 years worldwide in 2016 (Troeger et al., 2018).
More than 85% of the deaths happened in developing countries and less developed countries (Patton 2012). RV is divided into 8 groups (group A–H). The most common group A is divided into at least 27 G genotypes and 35 P genotypes. The most epidemic strains are P[4], P[6] and P[8] types (Matthijnssens et al. 2011). The host susceptibility to specific RV strains and the pathogenesis are influenced by genetically controlled expression of different histo-blood group antigens (HBGAs) among the world's population (Hu et al. 2012). Host phenotype might impact vaccine efficacy by altering susceptibility to vaccination or RV diarrhea (Lee et al. 2018). Therefore, investigation of the interaction mechanism between virus and host cells will provide a theoretical basis for the development of a safe and effective RV prevention and control system.
RV is a member of the Reoviridae family. It is a nonencapsulated, segmented double-stranded RNA virus surrounded by three layers of capsid protein. The RV genome consists of 11 segmental genes encoding a total of 6 structural proteins (VP1–4, VP6, VP7) and 6 non-structural proteins (NSP1–6) (Nakagomi and Nakagomi 2009). VP4, an outer capsid protein, is cleaved by a protease to VP5* and VP8* (Patton et al. 1993), of which VP8* is mainly involved in receptor recognition (Méndez et al. 1999). Studies have shown that VP8* recognizes sialic acid, which helps the virion to anchor to the cell surface, thereby mediating virus invasion into host cells (Sugiyama et al. 2004). Therefore, VP8* is the primary mediator of interaction between RV and host cell surface receptors.
Viral receptors are the key for viral invasion of the host, and the invading of most viruses is mediated by specific cellular receptors (Fleming et al. 2011; Grove and Marsh 2011; Howley and Knipe 2001). Studies have shown that RV has specific cellular tropism, infecting only the intestinal cells of the small intestine villi in vivo, and can only effectively infect renal epithelial cells and intestinal epithelial cells in vitro (Fleming et al. 2011; Trask et al. 2010). The invasion process presents a sequential, specific, multi-stage interaction with multiple receptor molecules on the host cell surface (Baker and Prasad 2010). However, the interaction between RV and its receptors is ambiguous so far.
Previously, we have constructed a bacterial surface display system expressing cleavable protruding domain protein of human norovirus (HuNoVs) with the help of N-terminal domain of the ice nucleation protein (InaQn) (Niu et al. 2015). It demonstrated that the displayed protein of HuNoVs could specifically recognize and bind to the HBGAs from porcine gastric mucin (PGM). The proteinHBGAs complex could be released from the host cells after digesting and low speed centrifugation (Xu et al. 2017). It is easy for us to separate the receptor candidates from natural sources without the background noise of bacterial cells.
In order to verify the universality of the system and further confirm the receptor binding ability, the VP8* of domestic epidemic genotype G9P[8] RV was expressed and displayed on the surface of Escherichia coli (E. coli) BL21 in this study. The function of bacterial cell surface display system was evaluated by Western blot and Enzyme-Linked Immunosorbent Assays (ELISA).
-
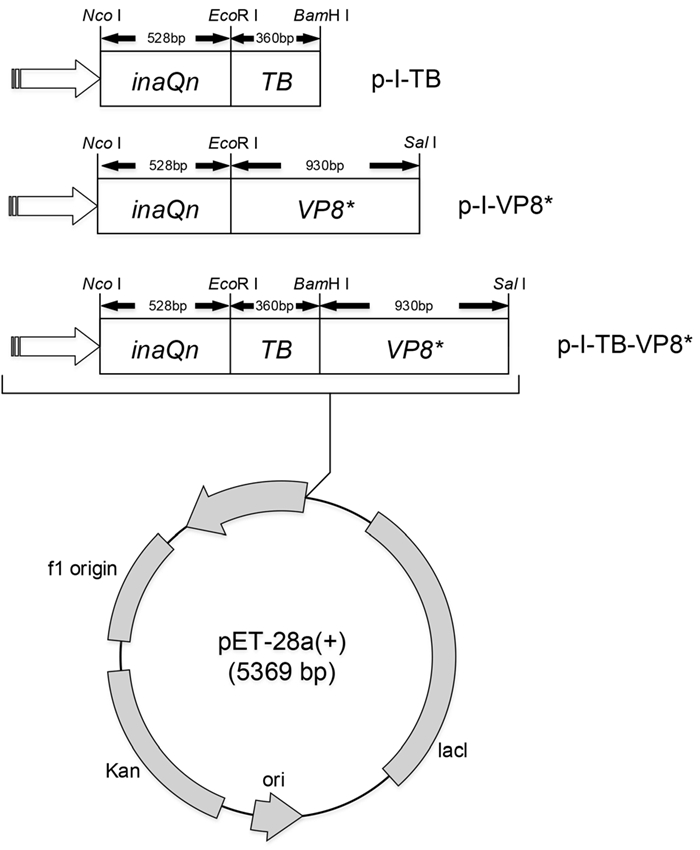
Competent E. coli DH5a and BL21 (DE3) (Thermo Fisher, Shanghai, China) were used for recombinant plasmid construction and protein expression, respectively. The nucleic acid fragment of a thrombin distinguished sequence "TB" was synthesized by Suzhou GENEWIZ Bio-Technology, Co., Ltd. Fragment of VP8* was kindly provided by Dr. Zhiyong Gao (Beijing Center for Diseases Prevention and Control). The inaQn, TB and VP8* nucleic acid fragments were inserted into the plasmid (pET-28a, ThermoFisher, Shanghai, China) respectively to make pET28a-inaQn-TB-VP8* (p-I-TB-VP8*). Similarly, the recombinant plasmid pET28a-inaQn-VP8* (p-I-VP8*) and pET28a-inaQn-TB (p-I-TB) were constructed and p-I-TB used as a negative control. Schematic diagram of recombined plasmids was shown in Fig. 1.
-
The recombinant bacteria were cultured in Luria-Bertani (LB, Sangon Biotech Co., Ltd., Shanghai, China) liquid medium containing 100.0 μg/mL kanamycin, at 37 ℃ shaken (150 rpm) overnight. The cultured bacteria were transferred to fresh LB medium (10.0 mL, 100.0 μg/mL kanamycin), and cultured at 37 ℃ with shaking (150 rpm) until OD600 reached 0.6. Isopropyl b-D-1-thiogalactopyranoside (IPTG; Merck, Germany) was added to a total concentration of 0.5 mmol/L, and incubated at 26 ℃ with shaking (120 rpm) for 12–16 h. The induced recombinant bacteria were adjusted to OD600 1.0 and stored at 4 ℃ for further use.
-
The prepared bacteria (100.0 mL) as described above were collected and washed twice with sterile PBS (pH 7.2), then resuspended in digestion buffer (1.0 mL, 20.0 mmol/L Tris-HCl and 150.0 mmol/L NaCl, pH 8.0). In accordance with manufacturer's guidelines for enzymatic activity, bovine thrombin (Yeason, Shanghai, China) was added at 1: 2000 to each digestion reaction and incubated at 37 ℃ for 3 h. The mixture was centrifuged at 4 ℃, 8000 ×g for 5 min. The protein was stored at - 80 ℃ for further use. The recombinant E. coli BL21 containing pET28a-inaQn-TB was treated in the same way as a negative control.
-
Recombinant E. coli BL21 strains containing constructed plasmids p-I-TB-VP8*, p-I-VP8* and p (pET-28a, negative control) were induced with IPTG and collected as described above. Surface-displayed VP8* was released from bacteria by thrombin digestion as described above. For SDS-PAGE, the IPTG-induced bacteria and the thrombinreleased VP8* were dissolved in 5 × SDS-PAGE loading buffer (Beyotime, Shanghai, China). Each sample was boiled for 5 min, and 10.0 μL of the sample was loaded and the samples were separated in a 12% SDS-PAGE gel, followed by staining with Coomassie Blue R250 (Beyotime, Shanghai, China). The polyclonal antibody against VP8* recombinant viral capsid protein (1:5000; kindly provided by Ningguo Feng at Stanford University) and peroxidase-conjugated goat anti-mouse IgG (H+L, 1: 3000; Yeasen, Shanghai, China) were used as primary and secondary antibodies in Western blot as described in our previous publication (Xu et al. 2017). The DAB Peroxidase Substrate Kit (Yeasen, Shanghai, China) was used as chromogenic substrate for Western blot.
-
After centrifugation, three groups of bacteria and supernatant obtained before and after thrombin digestion, were added to the wells (Sangon Biotech Co., Ltd., Shanghai, China) to incubate at 4 ℃ overnight for immune assay. Each well was washed 3 times with PBS, blocked with 120.0 μL of 1.0% BSA in PBS at 37 ℃ for 1 h, and then washed with PBS. One hundred microliters of polyclonal antibody against VP8* recombinant viral capsid protein was added to each well. Peroxidase-conjugated goat antimouse IgG (H+L chains, 1: 3000; Yeasen, Shanghai, China) was used as secondary antibody. All antibody incubation steps were performed at 37 ℃ for 1 h. The wells were washed 5 times with 120.0 μL of PBS-T after each incubation step. Then, 100.0 μL of 3, 3', 5, 5'-Tetramethylbenzidine (TMB, Frdbio, Wuhan, China) was added to each well. After incubating in the dark for 10 min, the chromogenic reaction was halted using 50.0 μL of 2 mol/L H2SO4. The OD450 values were measured by Sunrise Microplate Reader (Tecan Sunrise, Switzerland).
-
After IPTG induction, three groups of E. coli BL21 containing plasmid constructs p-I-TB-VP8*, p-I-TB and p, were collected by centrifugation, and then washed and resuspended in PBS as described above. The three groups of E. coli BL21 were incubated with 1.0 mg/mL (final concentration) type III PGM (Sigma, USA) and PBS at 37 ℃ for 1 h with gentle shaking, respectively. PBS was used as a negative control. Then bacteria were collected by centrifugation at 4 ℃, 8000 ×g for 5 min. After washing twice with PBS, the VP8* with captured ligands was released by thrombin digestion as described above. According to our previous reports (Xu et al. 2017; Niu et al. 2015), the optimal dilutions of anti-type A HBGA monoclonal antibody BG2 (MAb, Covance, Emeryville, CA, United States) and secondary antibody, a peroxidaseconjugated goat anti-mouse IgG (H+L chains; Yeasen, Shanghai, China) were determined experimentally to be 1: 1000 and 1: 3000 in blocking buffer. The steps of ELISA have been described above.
-
Saliva samples were collected from healthy people with different blood types, and treated according to previous reports (Nordgren et al. 2014; Wang et al. 2017). After being diluted in PBS with the ratio of 1: 100, the saliva samples were incubated with three groups of recombinant E. coli BL21 respectively as described above. PBS was used as a negative control. Then bacteria were collected by centrifugation at 4 ℃ at 8000 ×g for 5 min. The VP8* and captured ligands were released by thrombin digestion as described above. Interaction products or the treated saliva samples were added into immunoassay wells and incubated at 4 ℃ overnight. After blocking with 1.0% BSA, antiHBGAs monoclonal antibodies BG1-8 (Covance, Emeryville, CA, United States) were used as primary antibodies. A peroxidase-conjugated goat anti-mouse IgG (for BG 1, 2, 4, 5; Yeasen, Shanghai, China) or goat anti-mouse IgM (for BG 3, 6, 7, 8; Yeasen, Shanghai, China) were used as secondary antibodies. The steps of ELISA have been described above.
-
GraphPad Prism 7 software was used for statistical analysis. Each experiment was repeated at least three times (N > 3), which was independently repeated in triplicates (n = 3). Differences in means were considered significant when the P < 0.05, P < 0.03 (*), P < 0.002 (**), P < 0.001 (***).
Bacterial Strains and Construction of the Recombinant Plasmids
Culture and Expression of InaQn-TB-VP8* Fusion Proteins in E. coli BL21
Releasing of VP8* by Thrombin Digestion
SDS-PAGE and Western Blot
Validation of VP8* Expressed on Bacterial Surface of Recombinant E. coli BL21 by ELISA
Measuring the HBGAs-Binding Ability of Displayed VP8*
Measuring the Binding Ability of the System to Specific Receptors from Saliva Samples
Statistical Analysis
-
SDS-PAGE and Western blot were used to characterize the expression of the fusion proteins in bacteria and the thrombin-released VP8* (Fig. 2). The recombinant VP8* could be specifically recognized by its antibody. InaQnVP8*, InaQn-TB-VP8* fusion protein and thrombinreleased VP8* were approximately 55, 68 and 35 kDa, respectively. In Fig. 2, it demonstrated that the constructed VP8* was expressed successfully.
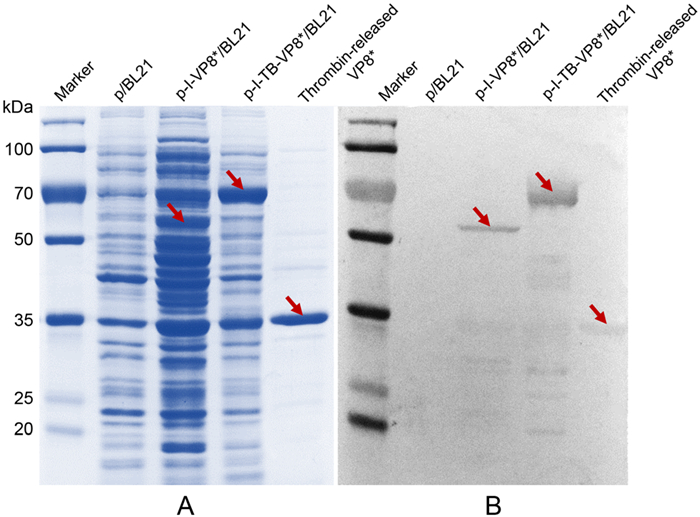
Figure 2. SDS-PAGE (A) and Western blot (B) analysis of the expression of E. coli cell surface display systems for VP8* of RV (G9P[8]). p/BL21 was a negative control. p-I-VP8*/BL21 and p-ITB-VP8*/BL21 were VP8* inserted cells without/with thrombin recognized sequence. Thrombin-released VP8* showed the VP8* protein that displayed on the surface of recombined cells and released by thrombin. Target proteins were marked by arrows.
-
In order to verify cell surface expression of the recombinant protein, the content of VP8* was detected in the supernatant after the enzyme digestion by ELISA. Samples were considered positive when the positive to negative (P/N) ratio was greater than 2.0. The results showed that the P/N value of the precipitated cells before digestion was more than 2.0, which indicated that VP8* was successfully expressed by E. coli BL21 (Fig. 3). In Fig. 3, the P/N value of the supernatant expressing VP8* after thrombin digestion was higher than that before digestion. The P/N value of the precipitated cells expressing VP8* before digestion was higher than that after enzyme digestion, which demonstrated that the VP8* were successfully anchored on the surface of E. coli cells. However, after digestion, the P/N value of the BL21 cells in the precipitate was still higher than 2.0, it might be due to the fact that a part of VP8* were expressed in the cell for the limitation of the surface sites of host bacteria. At the same time, the strains containing p-I-TB and p were used as negative controls in the whole process. There was no significant binding of VP8* antibody to the thrombin-digested expression product from negative control groups.
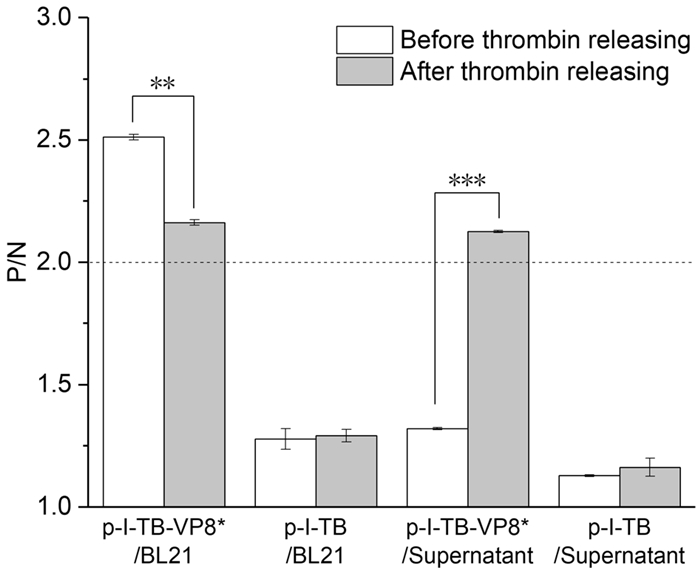
Figure 3. Detection of VP8* by ELISA before and after thrombin treatment. After thrombin treatment, VP8* was greatly reduced (**P < 0.002) in the cells and significantly increased (***P < 0.001) in the supernatant of p-I-TB-VP8*. There was no difference (P>0.05) observed in the cells and supernatant of p-I-TB. X-axis represents recombinant cells p-I-TB-VP8* and p-I-TB and their supernatant before and after thrombin treatment. The positive to negative (P/N) OD450 ratio is shown on the Y-axis. Negative controls included wells that contained all reagents but coated with PBS.
-
Three groups of recombinant E. coli BL21 containing p-ITB-VP8*, p-I-TB and p were incubated with PGM or PBS, respectively. The content of type A HBGAs in enzyme released supernatant was detected after thrombin digestion. The P/N value of incubation of p-I-TB-VP8*/BL21 with PGM was more than 2.0, which was significantly higher than that of others (P < .01), indicating that the detected HBGAs in the enzyme digested supernatant was derived from the incubated PGM instead of the bacteria themselves (Fig. 4). These results demonstrated that the VP8* expressed by the bacterial cell surface display system could specifically recognize and capture HBGAs from PGM.

Figure 4. Type A HBGA captured by displayed VP8* from porcine gastric mucin (PGM) after thrombin digestion. After incubating with PGM and then treated by thrombin, type A HBGA could be detected in the supernatant of p-I-TB-VP8* (***P < 0.001). X-axis represents three different recombinant cells used in this study. The positive to negative (P/N) OD450 ratio is shown on the Y-axis. Negative controls included wells that contained all reagents but coated with PBS.
-
Treated saliva from healthy people with different blood types was incubated with the bacterial cell surface display system of RV, and detected by ELISA. As shown in the Fig. 5, type A, B, Lewis a and Lewis b HBGAs were tested positive in saliva mixture with P/N ratios of 13.11, 2.30, 10.00, and 9.11, respectively. Results showed that type A HBGA was detected positive in the supernatant after sample incubation and thrombin digestion, indicating that VP8* expressed on the surface of E. coli could successfully recognize, capture and separate the type A HBGA from complex matrix. It demonstrated that the surface-displayed VP8* has the ability to separate specific conjugates from complex sample.

Figure 5. Capture ability of displayed VP8* to HBGAs in saliva mixture. Type A, B, Lewis a and Lewis b HBGAs were tested positive in saliva mixture (***P < 0.001). Only type A HBGA was tested positive by p-I-TB-VP8* (***P < 0.001). X-axis represents monoclonal antibodies specific for the HBGAs. The positive to negative (P/N) OD450 ratio is shown on the Y-axis. Negative controls included wells that contained all reagents but coated with PBS.
Characterization of Thrombin-Released VP8* Protein
Validation of Bacterial Cell Surface Expressed VP8* by ELISA
Specific Binding Ability of VP8* to Type A HBGA from PGM
Specific Binding Ability of VP8* to HBGAs from Mixed Saliva Sample
-
The target protein could be expressed on the surface of the bacteria with the help of InaQn, which has been well documented to be responsible for bacterial transmembrane and membrane-anchoring activity (Li et al. 2010, 2012). Previously, the cell surface display system using InaQn has been used in vaccine development. For example, a recombinant oral vaccine for hepatitis C has been tested (Lee et al. 2000). In our previous studies, the protruding domain protein of HuNoVs has been used to construct a bacterial surface display system, which has been proved to possess the ability to combine HBGAs, the well-accepted receptor from PGM (Niu et al. 2015). The bacterial surface display system could interact with HBGAs-like molecules from romaine lettuce (Wang et al. 2017). Furthermore, after updating the system, the receptor-P-protein complex could be released and isolated from bacteria surface by thrombin treatment, followed by low-speed centrifugation. The specific substance could be further analyzed without the background noise of expression host (Xu et al. 2017).
In order to verify the universality of this system, P protein of HuNoVs was replaced by VP8* in the bacterial surface display system in this study. The SDS-PAGE and Western blot results demonstrated that the fusion protein of VP8* was successfully expressed and anchored on the surface of E. coli. In Fig. 2, there were some background bands in SDS-PAGE, which were the bacterial proteins. In addition, little target protein may be broken when they were treated with boiled water bath before SDS-PAGE. The broken protein fragment would be recognized by the polyclonal antibody in Western bloting (Fig. 2). On the other hand, the increasing signal of supernatant after digestion indicated that the VP8* did be displayed on the surface of host cells and released easily by thrombin treatment with antigenicity which could be recognized by its specific antibody. The successful expression of VP8* illustrated that the bacterial surface display system could be widely used for a variety of exogenous protein expression and purification.
Receptors of RVs have been well characterized, including HBGAs (Hu et al. 2012), sialic acids (Yolken et al. 1987), heat shock cognate protein 70 (López and Arias 2004; Zarate et al. 2003), gangliosides (Guo et al. 1999; Iša et al. 2004) and integrins (Hewish et al. 2000; Graham et al. 2003). HBGAs play important roles in RV infection. Through the saliva- and oligosaccharide-based binding assays, HBGAs were recognized by RV VP8*, which was genotype-dependent (Huang et al. 2012). HBGAs can also be used as a polysaccharide receptor of some pathogenic microorganisms such as HuNoVs, RV and Helicobacter Pylori, which are related to the susceptibility of some infectious diseases (Harrington et al. 2004; Huang et al. 2012; Ilver et al. 1998). Barbe´ et al. (2018) reported that most VP8* from P[8] strains are more likely to bind to type Lewis b HBGA than to type A HBGA. However, other researchers also demonstrated that Lewis b and H type-1 HBGA were not involved in the recognition of common human rotaviruses (Böhm et al. 2015). Therefore, the exactly interactions between different genotypes of rotavirus and HBGAs are still unclear. Our lab is focusing on the study of the ligands/receptors that involved in the specially recognition of HuNoVs. We proposed that the substances that recognized by MAbs of HBGAs were some HBGAs-like substances instead of HBGAs. We have identified a HBGAs-like oligosaccharide (m/z: 1042, H2N2F2), separated from romaine lettuce extract and PGM, was involved in specific binding to HuNoVs particles with the help of bacterial surface display system. The structure of this oligosaccharide is a chimera of type A and Lewis a HBGA, which also contained the active structure of type H HBGA. We have demonstrated a completely new theory that the receptor for HuNoVs is not limited to HBGAs, but is indeed composed of HBGAs-like similar composition (data under review).
Saliva mixture with type A, B, Lewis a and Lewis b HBGA was utilized in this study to verify the specific recognition and combination of the RV bacterial surface display system (Fig. 5). After incubation and treatment by thrombin, the VP8*-receptor complex could be easily collected by low-speed centrifugation. The thrombin-released complex could be recognized by MAb (BG2) for type A HBGAs. It is consistent with the results we reported before that HuNoVs bacterial surface display system was able to capture and easily isolate viral receptors. The capacity to specifically enrich and purify receptors/ligands of expressed protein with this system has been further verified. To make clear the composition of the receptors/ ligands (including HBGAs), we are excavating the viral receptors from the MA 104 cell lines by using the developed VP8* cell surface display system. We believe that this system can be applied to other viral proteins and it is helpful for the discovery and identification of receptors of other pathogens.
-
This work was jointly supported by the National Key Research and Development Program of China (2017YFF0210200) and the National Natural Science Foundation of China (31772078).
-
DL and DW performed the experiments, DL, HG, ZZ and DW analyzed results and wrote the manuscript. DL, HG, ZZ, YX, DY and DW contributed to experimental design and carried out part of the experiments. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.














 DownLoad:
DownLoad: