HTML
-
Human cytomegalovirus (HCMV) is a beta-herpesvirus that can infect many kinds of human cells, including fibroblasts, endothelial cells, neuronal cells, monocytes/macrophages, and dendritic cells (Sinzger et al. 2008). HCMV often causes asymptomatic infection in healthy individuals; however, HCMV is a life-threaten pathogen in congenitally infected newborns and immunocompromised populations (Pass et al. 1986; Khoshnevis and Tyring 2002). To explore the pathogenesis of HCMV infection, many works that focused on the viral protein are done in the past. It's reported that HCMV IE1 protein promotes viral growth by impairing the antiviral function of SP100 proteins (Kim et al. 2011; Tavalai et al. 2011; Wagenknecht et al. 2015) and viral proteins UL97 and pUL83 (pp65) are beneficial to viral replication by disturbing the role of restriction factors include nuclear proteins IFN-γ inducible protein 16 (IFI16) (Landolfo et al. 2016). Recently, it is demonstrated that long non-coding RNA (lncRNA) RNA 4.9 participates in the establishment of HCMV latent infection in CD14(+) monocytes (Rossetto et al. 2013). However, the precise pathogenesis of HCMV infection is still unclear, and the relation between circRNAs and HCMV infection has rarely been reported until now.
Circular RNAs (CircRNAs) are a type of non-coding RNAs with a circular structure in which the 5' and 3' terminus are covalently linked together (Memczak et al. 2013). CircRNAs were firstly discovered in plant viruses in 1976 (Sanger et al. 1976). Nevertheless, circRNAs were regarded as a byproduct of the RNA splicing process concerned with their low abundance and unclear functions for a long time (Nigro et al. 1991; Capel et al. 1993; Cocquerelle et al. 1993). Gradually, more and more evidence revealed that circRNAs exist extensively and play crucial roles in many biological processes in organisms, including serving as miRNA or protein sponges (Salzman et al. 2012; Jeck et al. 2013; Memczak et al. 2013) and protein scaffolding (Du et al. 2016, 2017). Research supports that circRNAs regulate protein functions by serving as protein sponges or decoys. CircANRIL triggers nucleolar stress and activation of p53 in human vascular smooth muscle cells and macrophages by sponging pescadillo homologue 1 (PES1) (Holdt et al. 2016). Also, it's indicated that the synthase activity of cyclic GMP-AMP (cGAMP) synthase (cGAS) is blocked by circRNA ciacGAS, which decoys cGAS in dormant long-term hematopoietic stem cells (LT-HSCs) (Xia et al. 2018). In recent years, some studies are focused on the relationship between circRNAs and herpes virus infections. It was reported that Kaposi's sarcoma herpesvirus (KSHV) infection significantly changed host circRNA transcriptions in primary human umbilical vein endothelial cells (HUVECs) and MC116 (a KSHV-infected B cell line). Those influenced host circRNAs could repress viral gene expressions during infection (Tagawa et al. 2018). CircBARTs produced by Epstein-Barr virus, are highly expressed in infected tissue and cell lines with potential functions in viral oncogenesis (Toptan et al. 2018). Nonetheless, little is known about the relationship between host circRNAs and HCMV productive infection.
In our study, RNA sequencing (RNA-seq) was performed to investigate the circRNA profiles of mockinfected and HCMV-infected human embryonic lung fibroblasts (HELFs). Host circRNAs significantly influenced by HCMV productive infection were compared and analyzed. Transcriptions and sequences of noteworthy circRNAs (circSP100, circMAP3K1, circPLEKHM1, and circTRIO) were validated. Characteristics of circSP100 in HCMV-infected cells were further confirmed. Moreover, circSP100 potentially binding proteins were purified, determined, and analyzed, indicating multiple functions of circSP100 during HCMV productive infection.
-
The HELF cell line was acquired from the Shanghai Institute for Biological Sciences, Chinese Academy of Sciences (CAS). HELFs were cultured in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 units/mL streptomycin.
HCMV low-passage strain HAN (GenBank accession number: KJ426589.1) was isolated from a urine sample of a 5-month-old infant hospitalized in Shengjing Hospital of China Medical University (Zhao et al. 2016). 1 × 107 HELFs were prepared in 10-cm plates and infected with HAN at a multiplicity of infection (MOI) of 3.
-
HCMV infected HELF cells were harvested at 72 h postinfection (hpi). PBS-treated cells were harvested as control. Total RNAs were extracted using TRIzol Reagent (Thermo Fisher) according to the manufactures' instruction. The quality of the total RNAs was estimated using the NanoDrop ND-1000 spectrophotometer and Agilent Bioanalyzer 2100 (Agilent Technologies). Total RNAs were then subjected to ribosomal RNA depletion using Ribo-Zero rRNA Removal Kits (Epicentre). Linear RNAs were removed by RNase R treatment (Epicentre) according to the manufactures' instruction.
-
RNA fragmentation was performed randomly, resulting in fragmented RNA ranging from approximately 200 nucleotides (nt) to 500 nt. Fragmented RNA was reversely transcripted into cDNA using SuperScriptTM Ⅱ Reverse Transcriptase (ThermoFisher) according to its protocol. Libraries were constructed using the TruSeq RNA Sample Prep Kit (Illumina). Paired-end RNA sequencing was performed on IlluminaHiSeq 2500 at Biotechnology Co. Ltd., Shanghai, China. The RNA sequencing data have been deposited in the Gene Expression Omnibus database under accession number GSE138836.
-
Ribosome RNA reads and reads less than 25 nt or containing adaptor sequences were filtered using Seqtk (https://github.com/lh3/seqtk). Clean reads were mapped to reference genomes (GRCH37/hg19) in the UCSC genome browser (http://genome.ucsc.edu/). As described previously (Li and Durbin 2010), the unmapped reads were used for further identification. Sequences from both ends of the unmapped reads were extracted and aligned independently to find unique anchor positions within spliced exons by CIRI (Gao et al. 2015). CircRNAs splicings were predicted by anchors that aligned in the reversed orientation (head-to-tail).
The predicted circRNAs were aligned with circBase (http://circrna.org/). Using ANNOVAR database (Wang et al. 2010), splicing ends of each circRNA candidate were mapped to the genomic regions, and then the results were compared to the RefSeq and UCSC databases to annotate the functional element of circRNA.
-
Junction reads for each circRNA candidate was normalized to spliced reads per billion mappings (SRPBM) to compute the circRNA expression (Li et al. 2015). Differential circRNA transcriptions were calculated according to SRPBM with edgeR (Robinson et al. 2010). The q-value (adjusted P value) was achieved by controlling the false discovery rate (FDR). Compared to circRNAs in mock-infected cells, circRNAs with > 2 (or < 0.5) fold changes and q-value < 0.05 were determined to be significantly differential circRNAs.
To assess the potential biological function of differential circRNAs, we performed a cluster analysis of the parental genes of differential circRNAs for Gene Ontology (GO, http://www.geneontology.org/) and KEGG (https://www.kegg.jp/).
-
2 μg of total RNA was subjected to RNase R treatment (5 units/μg RNA) at 37 ℃ for 30 min and reversely transcribed using PrimeScriptTM RT-PCR Kit (TaKaRa). To verify the PCR specificity for circRNAs, we amplified cDNAs of linear SP100 and linear GAPDH as RNase R non-resistant controls. Divergent primers were designed to target circRNA junction sites, while convergent primers were designed to target linear SP100 (NM_001080391) and linear GAPDH (NM_002046.7). Primer information is listed in Supplementary Table S1. 100 ng of cDNA was amplified using the PrimeScriptTM RT-PCR Kit (TaKaRa) according to the manufacturer's protocol. PCR products were analyzed by electrophoresis on 1.5% agarose gel and visualized under UV-light. PCR products with expected sizes were purified using WizardⓇ Plus SV Minipreps DNA Purification System (Promega) and cloned into pCR 2.1 vector using TA CloningTM Kit (Thermo Fisher). Sequencing was performed using Sanger sequencing. Sequences were analyzed using DNAClub and BioEdit software.
-
CircSP100 was detected with sense probes and antisense probes. The sense probes target antisense transcripts, while antisense probes target sense transcripts. The specific probes were synthesized and labeled in vitro with DIGUTP (Roche) using DIG Northern Starter Kit (Roche) according to manufacturer's protocol. Primers for generating these probes are listed in Supplementary Table S1.
20 μg of RNA was separated by electrophoresis on agarose gel with 2% formaldehyde and transferred to IMMOBILON-NY+ nylon membranes (MILLIPORE) by capillary transfer. Subsequently, the membrane was rinsed in 2 × SSC for twice briefly and baked at 80 ℃ for 2 h. The membrane was hybridized with 2 μL of DIG-labelled probes at 60 ℃ overnight. Stringency wash in 2 × SSC and 0.1% SDS at room temperature was performed twice, followed by a rinse in 2 × SSC and 0.1% SDS at 68 ℃ twice. The membrane was then incubated with anti-DIGAP. Specific detection was visualized with the addition of CDP-Star and imaged by Bio-Rad molecular imager chemiDox XRS with ImageLab software (Bio-Rad). Quantities of 28S and 18S rRNAs in each sample were used as the loading controls.
-
2 μg of total RNA was subjected to RNase R treatment (5 units/μg RNA) at 37 ℃ for 30 min and reversely transcribed using the PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa). Quantitative PCR (qPCR) was performed on Applied Biosystems 7300 using TB Green Premix Ex TaqTMⅡ (TliRNase H Plus) (TaKaRa) according to the manufacturer's protocol. Primer information for circSP100, linear SP100, and linear GAPDH amplification are listed in Supplementary Table S1. Each sample was performed in three replicates. For the kinetic of circSP100 and linear SP100, the result was shown using 2-ΔΔCT value.
-
5 × 106 HCMV-infected HELF cells (72hpi) were collected for subcellular fractionations. Nuclear and cytoplasmic fractionations were performed using the Cytoplasmic & Nuclear RNA Purification Kit (NORGEN) according to the manufacturer's protocol. Semiquantitative PCR of circSP100 was performed in cytoplasmic and nuclear RNA, respectively. Human U2 small nuclear 1 (RNU2-1, NR_002716) and GAPDH were amplified as the nuclear and cytoplasmic reference genes, respectively. The primers are listed in Supplementary Table S1.
-
RNA probe library containing 11 probes antisense to sequence around the circSP100 junction site was used for RNA antisense purification (RAP). RNA probes were designed to be 120 nt in length, imbricating the junction site (Supplementary Fig. S1A). T7 promoter sequence was added to the 5' terminus. DNA sequences of probes before in vitro transcription were amplified by RT-PCR, and the primers are listed in Supplementary Table S2. Biotinlabeled RNA probes were produced using HiScribe T7 High Yield RNA Synthesis Kit (NEB) and Biotin-16-UTP (Roche) according to the manufacturer's instruction.
-
Batches of 0.5 × 108 HCMV-infected HELFs (72hpi) were washed once with iced PBS and subjected to crosslink on ice using Spectrolinker UV Crosslinker (8000 J/cm2 of UV at 254 nm). Cells were collected by centrifugation at 2000g for 5 min at 4 ℃. Meantime, HCMV-infected HELFs without crosslinking were collected as the control for the non-specific binding.
The total cell lysate preparation and RAP were performed was as previously described (McHugh et al. 2015) with modification. The change included in our study was as followed: ultrasonication was omitted to maintain the intact circRNA structure in cell lysate preparation procedure. Biotinylated RNA probes were used for the hybridization, and 100 μL of Streptavidin DynabeadsMyOne C1 magnetic beads (Thermo Fisher) were used for each sample. Incubation of probes/lysate mixtures and beads/probes/lysate complexes were performed at 45 ℃ with shaking in a shaking bath. At the end of the elution, samples were stored at - 80 ℃ for further analysis.
RNA of the elution sample and RNA input sample were magnetically separated and incubated with 1 mg/mL Proteinase K (Beyotime) at 55 ℃ for 1 h. Then, RNA was recovered using SILANE beads (ThermoFisher) according to the manufacturer's instruction. RT-PCR was performed as previously mentioned. Amplification of circSP100 sequence, including junction site, indicated the specific purification of circSP100 and amplification of linear GAPDH was used to monitor the non-specific purification. Here, we also amplified the cDNA transcribed from the circSP100 RAP RNA probes, which referred to the negative control for the RT-PCR.
-
SDS-PAGE gels were prepared using the SDS-PAGE reagent kit (Beyotime). Each of 8 μL of RNA elution sample (non-crosslinked and UV crosslink groups) was mixed with 2 μL of 5 × SDS-PAGE Sample Loading Buffer (Beyotime) and heated at 100 ℃ for 10 min, followed by ice incubation for 10 min. Electrophoresis was performed at 50 V for concentration and ran at 100 V for 1.5 h during the separation. Then, silver staining was performed using Protein Stains K Kit (Sangon Biotech) according to the manufacturer's protocols.
-
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was conducted for each of 20 μL of RAP final products (non-crosslinked and UV crosslink groups) as previously described (Kalli and Hess 2012; McHugh et al. 2015). Concisely, liquid chromatography was performed on an EASY-nLC 1000 UPLC system. The separation was performed at a constant flow rate of 350 nL/min. The gradient was set as follows: 6%–23% solvent B (16 min), 23%–35% solvent B (8 min), 40%–80% solvent B (4 min), and 80% solvent B (4 min). The peptides were then subjected to nanospray ionization (NSI) at 2.0 kV. Tandem mass spectrometry (MS/MS) was operated in data-dependent acquisition (DDA) mode to alternate between a full scan (m/z = 350–1800) in an Orbitrap Q Exactive Plus mass spectrometer (Thermo). Higher energy collision dissociation (HCD) tandem mass spectrometry scans (resolution = 17, 500) were performed after the full scan (resolution = 70, 000) on top 20 most abundant ions using automatic gain control (AGC) of 1 × 105 ions. The maximum ion fill-time for MS/MS scans was 100 ms, the HCD-normalized collision energy (NCE) was 28, and the dynamic exclusion time was 15 s.
The resulting data were processed using Proteome Discoverer 1.3, and tandem mass spectra were searched against homo sapiens and human cytomegalovirus entries database (20, 590 entries in total). Trypsin was specified as a cleavage enzyme allowing up to 2 missing cleavages. The mass error was set to 10 p.p.m for precursor ions and 0.02 Da for fragment ions. Carbamidomethyl on Cys was specified as a fixed modification, and oxidation on Met was specified as variable modification. Peptide confidence was set at high, and peptide ion score was set > 20.
-
All statistics were calculated using Prism Version 8.0 (GraphPad, San Diego, CA 92108, USA). The data in RT-qPCR are presented as mean ± standard deviation (SD).
Cell Culture and HCMV Infection
RNA Extraction
RNA Sequencing
circRNAs Prediction
circRNAs Data Analysis
RT-PCR and Sequencing
Northern Blot
RT-Quantitative PCR (RT-qPCR)
Subcellular Fractionation
RNA Probe Preparation
RNA Antisense Purification
Silver Staining
Mass Spectrometry
Statistic Analysis
-
In our study, a total number of 27, 409 and 35, 515 distinct host circRNAs were identified in mock-infected and HCMV-infected HELFs, respectively (Fig. 1A). The host circRNA profiles in mock-infected and HCMV-infected HELF cells shared a similar pattern in the distribution of backspliced read accounts. More than 96% of the backspliced read accounts of the identified host circRNAs were less than 100 (Fig. 1A and Supplementary Table S3). Referring to the issues where the circRNAs originate from, it was found that more than 82% of host circRNAs were derived from exons; the others were obtained from introns or intergenic regions.

Figure 1. Host circRNAs profiles in mock-infected and HCMV-infected HELFs. A circRNAs expressed in mock-infected and HCMV-infected HELFs; B The distribution of circRNAs in the human chromosome between mock-infected and HCMV-infected HELFs; C The number of circRNAs derived from one gene. 8, 138 host genes produced 40, 274 kinds of circRNA in this study (intergenic circRNAs are excluded); D The number of circRNAs in this study which were annotated in circBase. 15, 798 of 43, 200 circRNAs overlapped to circBase.
Generally, host circRNAs were transcribed abundantly and diversely in HELF cells. By analyzing the genomic locations of the parental genes of circRNAs, it was found that the host circRNAs were transcribed extensively throughout the genomes, and no difference of the distribution was found between mock-infected and HCMV-infected cells (Fig. 1B). Alternative splicing often occurs during mRNA transcription. In our results, alternative splicing was also observed for circRNA transcription. Excluding intergeniccircRNAs, about 70% of circRNAs have more than two various isoforms of circRNAs (Fig. 1C). In total, 40, 274 circRNA variants were produced from 8, 138 host genes due to alternative splicing. By comparison, approximately 37% of circRNAs identified in this study (15, 798 of 43, 200) were annotated in circBase (Fig. 1D). The remainder was novel circRNAs.
-
CircRNAs profiles of mock-infected and HCMV-infected HELFs were compared to address the effects of HCMV infection on circRNA transcriptions. Generally, 19, 724 host circRNAs were found in both mock-infected and HCMV-infected cells (Fig. 2A), 15, 791 host circRNAs newly appeared after infection, and 7685 circRNAs were found only in mock-infected cells. By selecting circRNAs of which fold-change > 2 (or < 0.5) and q-value < 0.05, 283 host circRNAs were identified to be significantly changed by HCMV infection (Fig. 2B). Detailed information is listed in Supplementary Table S3. Among them, transcriptions of 184 host circRNAs were up-regulated significantly after infection, whereas transcriptions of 99 circRNAs were down-regulated (Fig. 2B).
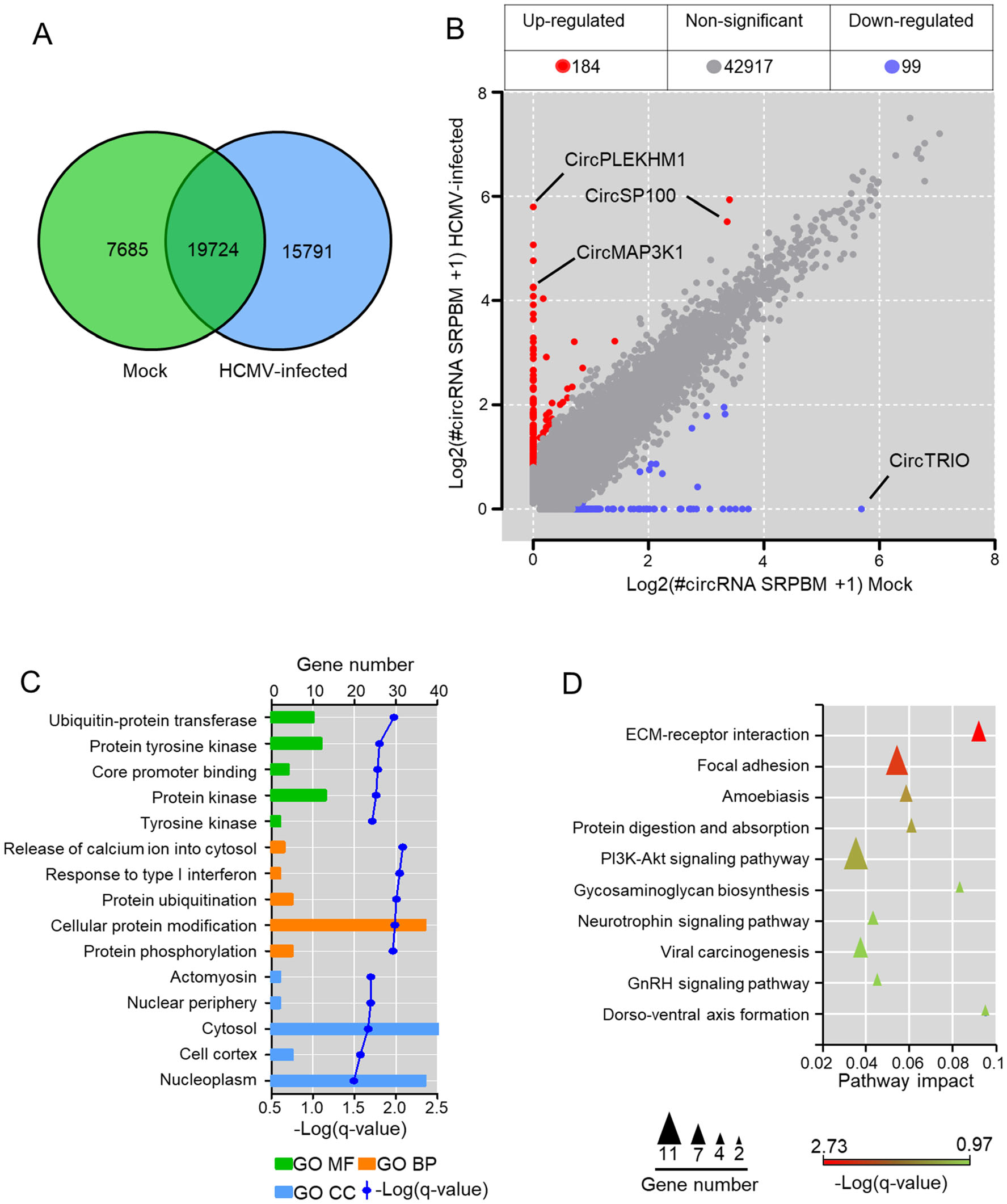
Figure 2. Alterations of circRNA transcriptions by HCMV infection in HELFs. A Venn diagrams of differentially expressed circRNAs in RNA-seq; B circRNAs significant differential based on standards of > 2 (or < 0.5) fold-change and q-value < 0.05 between mockinfected and HCMV infected HELFs; C GO enrichment of host genes of the significant change circRNAs. Only the top five of each category (molecular function, biological process, and cellular component), which were arranged by q-value, are presented; D KEGG enrichment of host genes of the significant change circRNAs. The top ten of the pathway arranged by q-value is exhibited.
To investigate the potential functions of the differentially expressed circRNAs associated with HCMV infection, the parental genes of these changed circRNAs were assigned to GO term analysis with Gene Ontology. Consequently, the parental genes were enriched among 44 biological processes, 16 molecular functions, and nine cellular components, all of which were filtered by the threshold value of q-value < 0.05. The top five enrichment of each GO term analysis is presented in Fig. 2C. The parental genes were mainly enriched in cellular processes involved in the release of calcium ion into the cytosol, response to type I interferon, protein ubiquitination, cellular protein modification, and protein phosphorylation. Meanwhile, KEGG enrichment was also performed for parental genes of differential circRNAs. The parental genes were indicated to be enriched in pathways involved in ECM-receptor interaction, focal adhesion, amoebiasis, and PI3K-Akt signaling pathway (Fig. 2D). The results suggested that circRNAs significantly changed by HCMV infection might play potential roles in the regulation of viral entry, apoptosis, and cell proliferation.
-
Transcription levels of host circRNAs significantly changed by HCMV infection were shown in the heat map (Fig. 3A). Candidate circRNAs with significant differences (circPLEKHM1 and circTRIO) or attractive known functions for their parental genes (circSP100 and circMAP3K1) were selected for further validation by RT-PCR using RNA post-RNase R treatment. To verify the PCR specificity for circRNAs, we amplified cDNAs of linear SP100 and linear GAPDH as RNase R non-resistant controls. As shown in Fig. 3B, since linear RNAs are unresistant to RNase R digestion, products of linear SP100 and GAPDH markedly decreased after RNase R treatment. No decrease was observed for circRNAs products after RNase R digestion.
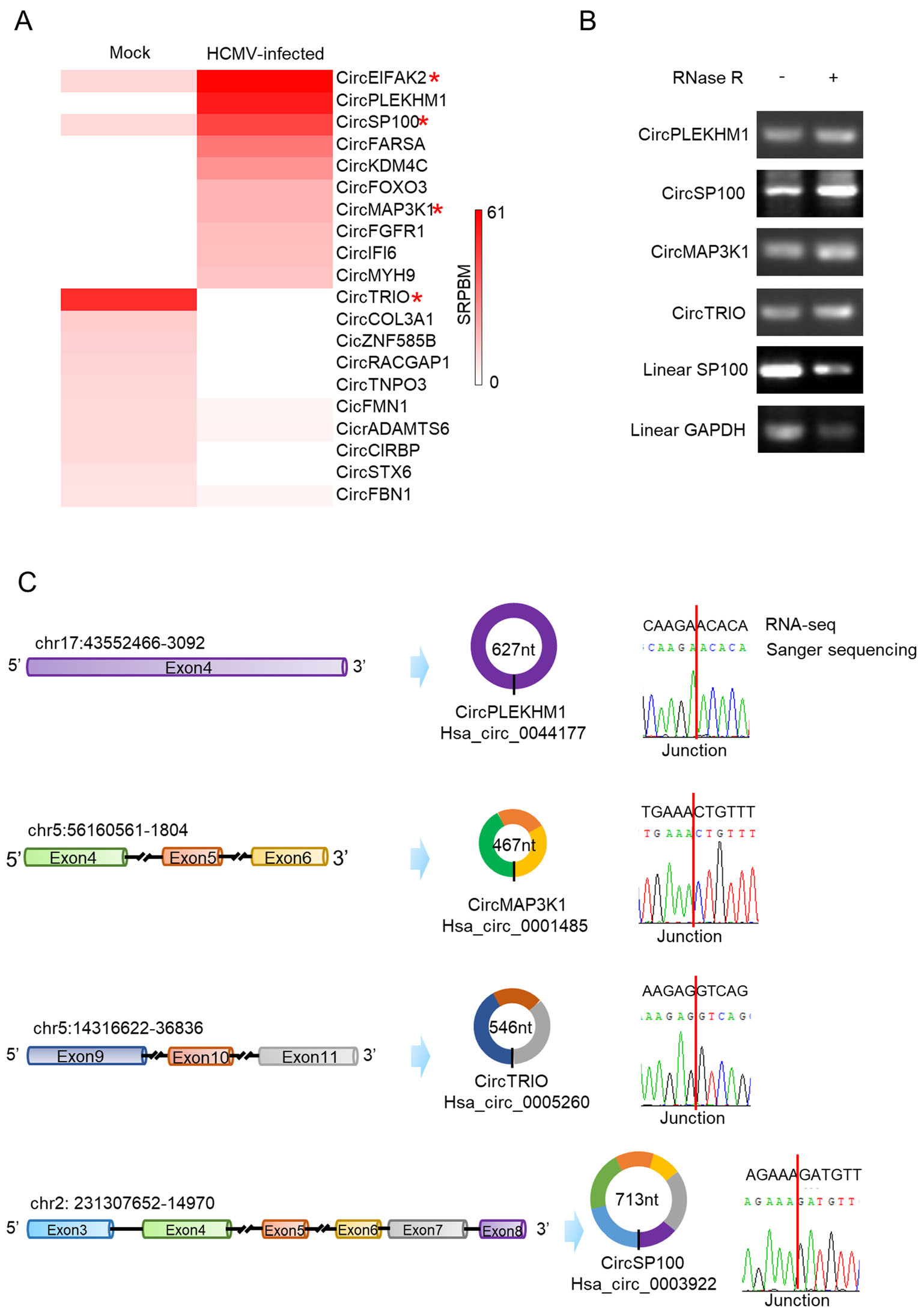
Figure 3. Validation of candidate circRNA transcriptions changed by HCMV productive infection. A Heatmap of the significant upregulated and down-regulated circRNAs. Red star symbols mark the circRNAs selected for further verification; B Verification of cyclization for the selected circRNAs by RNase R treatment. RNase R treatment indicates noticeable depletion of linear RNA (SP100 and GAPDH) but not affect the circRNAs (circPLEKHM1, circSP100, circMAP3K1, and circTRIO); C Verification of the selected circRNAs by Sanger sequencing. Sanger sequencing indicated all of the chosen circRNAs were consistent with RNA-seq results.
PCR products were cloned and sequenced then. As indicated in Fig. 3C, the sequences of circRNA junction sites were consistent with our previous RNA-seq results and data in CircInteractome (Dudekula et al. 2016). The full sequence information of circSP100, circMAP3K1, circPLEKHM1, and circTRIO was achieved by employing the "rolling" replication strategy during the PCR process (Barrett et al. 2015; You et al. 2015). By alignment with their transcripts, it was confirmed that the validated circRNAs were exoniccircRNAs and consisted of multiple exons except for circPLEKHM1 (Fig. 3C).
-
Northern blot showed that circSP100 was a sense transcript as predicted and increased after infection (Fig. 4A). Subcellular fractionation and RT-PCR indicated that circSP100 was preferentially localized in the cytoplasm (Fig. 4B).
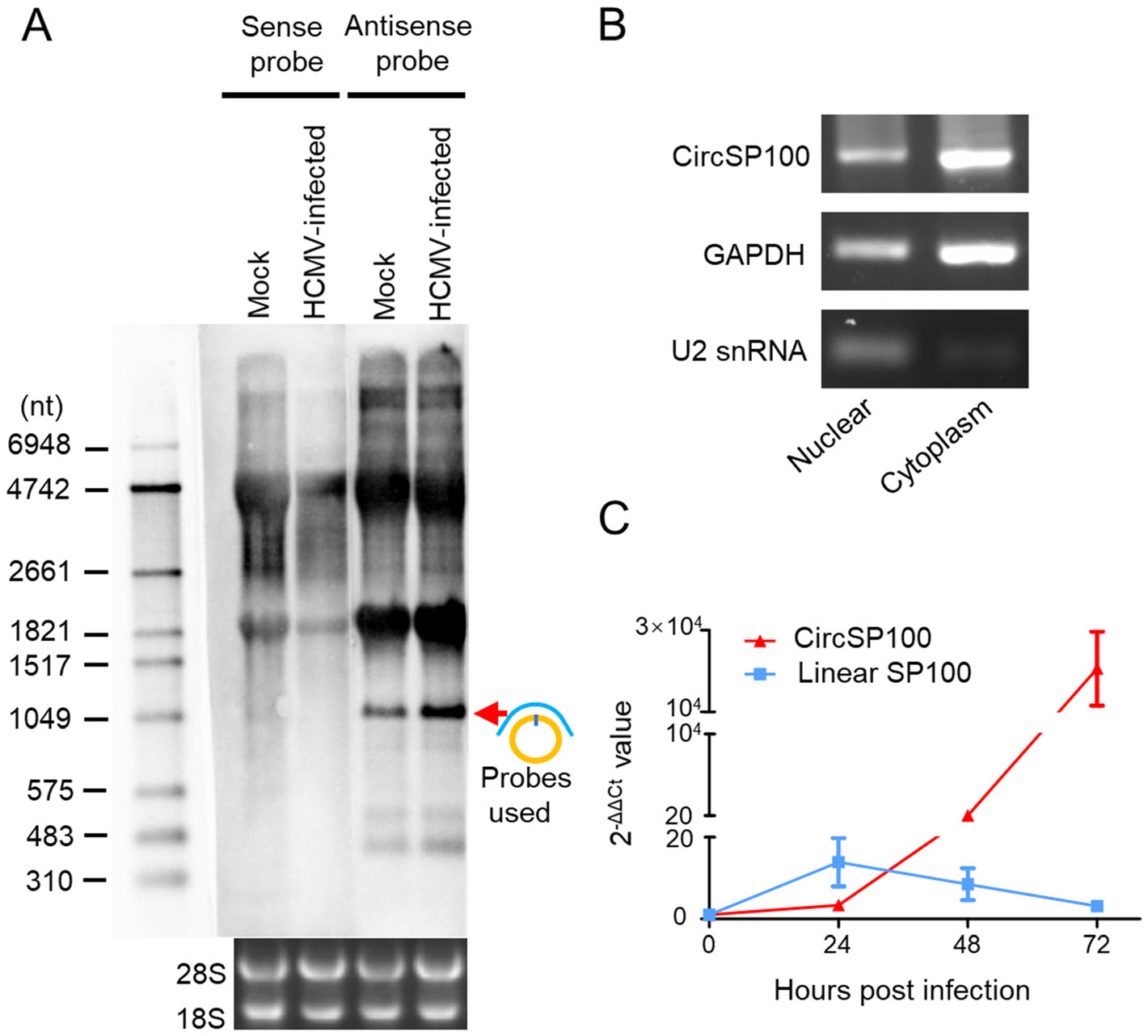
Figure 4. Characteristics of circSP100 during HCMV productive infection. A Northern blot of circSP100. The red arrow indicates the circSP100 RNA transcript; B Subcellular fractionation of circSP100. GAPDH and U2 snRNA were used as the markers for cytoplasm and nuclear fractionation, respectively; C RT-qPCR for the transcript level of circSP100 and linear SP100 mRNA (NM_001080391) in HELFs infected with HCMV at the suggested time points. The data represent the mean ± SD from three replicates.
As known, productions of linear RNA and circRNA maintain homeostasis at transcriptional levels (Ashwal-Fluss et al. 2014; Liang et al. 2017). To investigate the relationship between transcriptions of linear SP100 and circSP100, we analyzed the kinetics of SP100 mRNA, and circSP100 at different time points post-infection. It was found that circSP100 levels were increased continuously post-infection, while SP100 mRNA levels were increased before 24 hpi and then dropped gradually (Fig. 4C).
CircRNAs often serves as a scaffold or protein sponge (Holdt et al. 2016). For the preliminary investigation of circSP100 potential functions, RAP was conducted to pull down circSP100 binding proteins (Fig. 5A). The capture of circSP100 was confirmed by RT-PCR (Supplementary Fig. S1B). Proteins pulled down by RAP were detected by silver staining (Fig. 5B). Protein samples were analyzed by mass spectrometry.
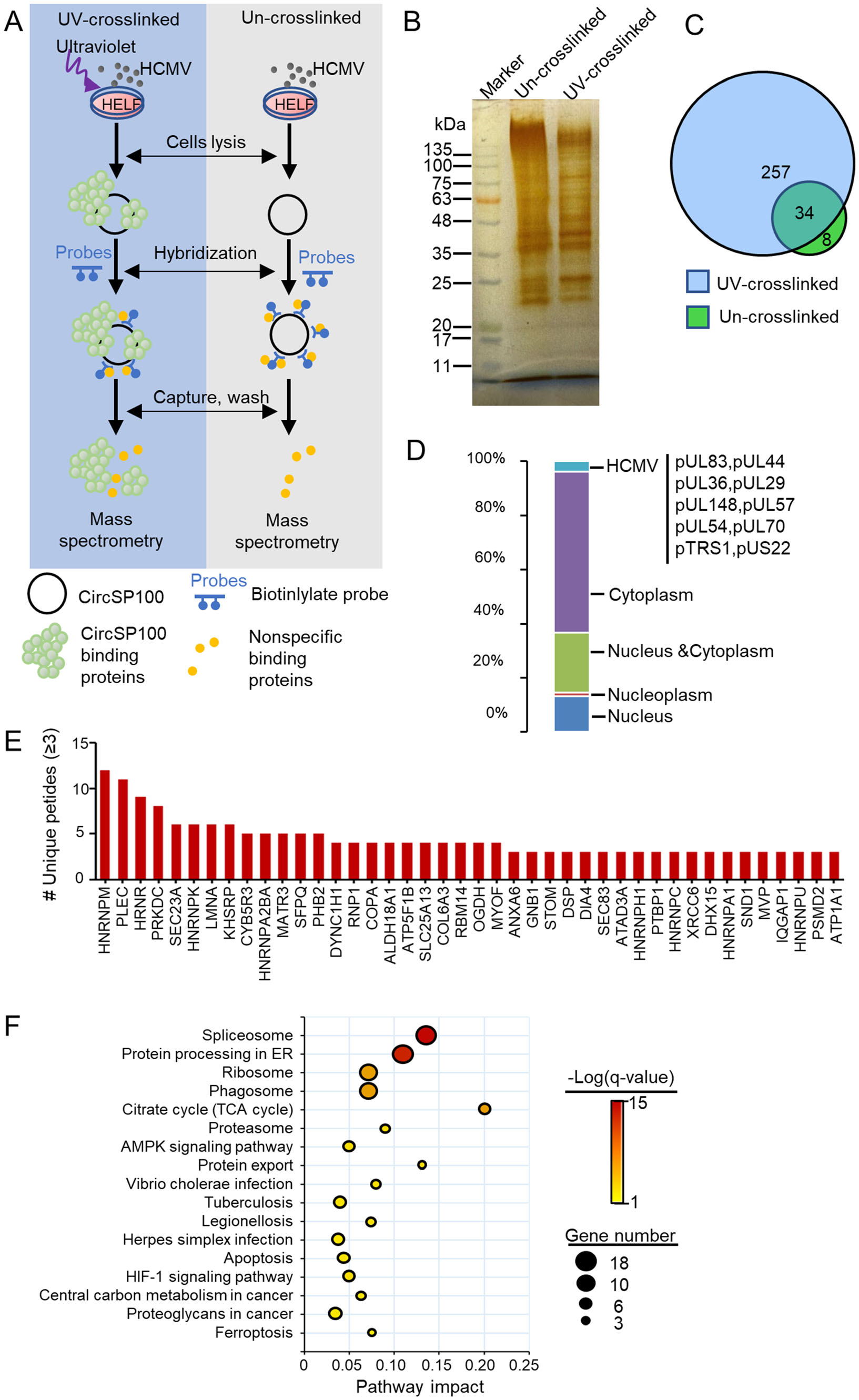
Figure 5. Analysis of circSP100 binding proteins captured by RAP. A The strategy of RAP mass spectrometry; B Proteins purified with RAP from UV-crosslinked or un-crosslinked lysis extracted from HCMV infected HELF cells; C Venn diagrams of proteins purified in RAP; D Subcellular location of the proteins that interacted directly with circSP100; E The circSP100 binding proteins. Only the proteins of unique peptides ≥ 3 were indicated; F KEGG enrichment of the proteins that interacted with circSP100.
In general, 291 proteins were recognized in samples from UV-crosslinked cells and 42 host proteins in samples from the control cells (Fig. 5C and Supplementary Table S4). Excluding the 34 proteins both in UV-crosslinked cells and control cells, 257 proteins were identified to be circSP100-binding proteins, among which 10 were HCMV encoding proteins. About 59% of the circSP100-binding proteins were determined to be cytosolic proteins, whereas 37% were nucleoproteins (Fig. 5D).
Captured proteins with top unique peptide values were listed in Fig. 5E. A total of 257 circSP100 binding proteins were analyzed for KEGG enrichment. The results suggested these circSP100-binding proteins were mainly involved in the spliceosome, protein processing in ER, ribosome, and phagosome pathways (Fig. 5F), indicating the multifunctional characteristic of circSP100 during infection. Notably, 18 of the circSP100-interacting proteins were involved in the spliceosome process (including HNRNPM, HNRNPK, DDX5, DHX15, HNRNPA1, HNRNPC, HNRNPU, PCBP1, SRSF1, SRSF6, SNRPB, SNRPD1, SNRPF, SART1, SNRNP200, SF3B3, TRA2A, and HNRNPA3). It is suggested that circSP100 might involve in spliceosome associated pathway by working as protein sponges for one or more of these 18 kinds of proteins. Moreover, PRKDC (also known as DNA-PKcs) and XRCC6 (also known as Ku70) were also identified as circSP100 interacting proteins with high unique peptide values in the RAP assay. PRKDC and XRCC6 are role components of DNA-dependent protein kinase (DNA-PK) complex that serves as a DNA sensor and induces innate immune response during DNA virus infection (Nussenzweig et al. 1996; Gu et al. 1997). It was speculated that circSP100 might participate in the formation of DNA-PK as a scaffold, or inhibit the formation of DNAPK by consuming PRKDC and XRCC6 as a protein sponge during HCMV infection.
Host circRNA Profiles in Mock-Infected and HCMV-Infected HELFs
Alterations of circRNA Transcriptions by HCMV Infection in HELFs
Validation of Candidate circRNA Transcriptions Changed by HCMV Productive Infection
Characteristics and Potential Functions of circSP100 during HCMV Productive Infection
-
HCMV is apt to cause abortive infection in healthy humans. It is estimated that the global seroprevalence of HCMV IgG antibodies is 83% in the general population, 86% in women of reproductive age, and 86% in donors of blood or organs (Zuhair et al. 2019). However, HCMV infection is fatal for the immunocompromised population and newborns (Pass et al. 1986; Khoshnevis and Tyring 2002). The pathogenesis of HCMV infection remains poorly understood. Many non-coding RNAs have been recently confirmed to be involved in the pathogenesis of HCMV infection, such as microRNAs (Guo et al. 2015; Jiang et al. 2017) and long non-coding RNAs (lncRNAs) (Mohammad et al. 2017). Nevertheless, the relationship between host circRNAs and HCMV infection is unclear.
It has been found that infection of herpesviruses could change host circRNA profiles. Host circRNAs changed by KSHV infection might serve as antiviral factors (Tagawa et al. 2018). Recently, it is found that HCMV latent infection could alter host circRNA transcriptions in THP-1 cells (Lou et al. 2019). In our study, circRNA profiles were analyzed in HELF cells with or without HCMV productive infection. 184 host circRNAs were up-regulated and 99 down-regulated post HCMV infection (Fig. 2B). The host circRNA profiles of mock-infected and HCMV-infected cells shared similar characteristics, such as the distribution of backspliced read accounts, the original of host circRNA, and the genomic location of the parental gene of circRNAs. KEGG enrichment showed that the host genes of differential circRNAs were related to ECM-receptor interaction, focal adhesion, amoebiasis, and PI3K-Akt signaling pathway (Fig. 2C and 2D). As known, the PI3K-Akt signaling pathway is activated by many types of cellular stimulation and increases metabolism, growth, proliferation, and cell survival, all of which are helpful to virus replication (Plas and Thompson 2005; Buchkovich et al. 2008). During HCMV infection, transient and consistent activation of PI3K-Akt was activated by the attachment between HCMV and viral receptors (Johnson et al. 2001), and expression of IE1 and IE2 proteins, respectively (Johnson et al. 2001; Buchkovich et al. 2008). Consequently, HCMV lytic replication occurs duce to the activation of the PI3K-Akt pathway (Johnson et al. 2001; Buchkovich et al. 2008). ECM-receptor and focal adhesion are reported to be associated with virus entry, spread, and viral persistence. Integrins, the most critical ECM-receptors in cells, were hijacked during viral infection (Hamidi and Ivaska 2018). It's reported that integrin α3β1 promoted KSHV entry into target cells by interacting with KSHV envelope glycoprotein B and inducing the integrin-dependent activation of focal adhesion kinase (Akula et al. 2002). Also, it's suggested that HCMV uses epidermal growth factor receptor (EGFR)- and integrin-dependent signaling pathways to activate signal transducer and activator of transcription (STAT) 1, which promotes the motility, differentiation, and polarization of the infected cell (Collins-McMillen et al. 2017). Furthermore, it's pointed out that focal adhesion damage facilitates HCMV infection in HCMV-infected cells (Stanton et al. 2007). Collectively, it's suggested that the host gene of the significantly changed circRNAs may be associated to HCMV infection in viral entry, replication, spread, and viral persistence.
Four host circRNAs with significant differences or attractive functions for parental genes were further focused: circPLEKHM1 was only found in HCMVinfected cells at the high transcriptional level, while circTRIO was oppositely found only in HELF cells and eliminated by HCMV productive infection; MAP3K1 is involved in signal transduction events, including the ERK/JNK kinase pathway (Xia et al. 1998; Ye et al. 2007) and NF-kappa-B pathway (Xia et al. 1998); SP100 plays vital roles in tumorigenesis (Held-Feindt et al. 2011), gene regulation (Yordy et al. 2004, 2005), and antiviral responses to HCMV infection (Kim et al. 2011; Tavalai et al. 2011; Wagenknecht et al. 2015; Landolfo et al. 2016). The transcriptions and sequences of these host circRNAs were then validated consistently to those results of RNA sequencing. It was confirmed that HCMV productive infection influences the transcription of host circRNAs. It was speculated that the circRNAs changed by HCMV productive infection might affect the expressions or functions of their parental genes associated with infection.
In our results, circSP100 is 713 nt in length and contains six exons (back-spliced at 5' terminus of exon 3 and 3' terminus of exon 8) (Fig. 3C). By northern blotting with sense and antisense probes, it was confirmed that circSP100 was transcribed from the antisense strand of the genome. It seemed that circSP100 might be produced from the same pre-RNA with SP100 mRNA. To address the relationship between circSP100 and its parental gene, the transcriptions of circSP100 and SP100 mRNA were compared at different time points post-infection. The results showed that circSP100 transcriptions increased along with HCMV infection, whereas SP100 mRNA decreased after 24hpi. It was speculated that since circSP100 and SP100 mRNA were produced from the same pre-RNA strand, the homeostasis between circSP100 and SP100 production might be broken upon HCMV infection. The more circSP100 was created, the less SP100 mRNA was transcribed. It would lead to a reduction of SP100 protein expression, benefiting HCMV survival.
CircSP100 was found mainly located in the cytoplasm, indicating it might play roles by binding with cytoplasmic molecules. Hence, we performed RAP to capture potential binding proteins of circSP100. Consequently, 257 proteins were detected binding to circSP100 (Fig. 5C). These proteins involved in multiple bioprocesses, such as spliceosome, protein processing in ER, ribosome, and phagosome. It has been demonstrated that inhibition of canonical mRNA splicing by depleting core spliceosomal components could increase circRNA transcriptions (Liang et al. 2017). Eighteen proteins involved in the spliceosome process were found in our RAP results. HCMV infection may induce the interaction between circSP100 and one or more of these eighteen kinds of proteins, resulting in the dysfunction of the spliceosome. Subsequently, the balance of transcription shifted from SP100 mRNA to circSP100, result in the reduction of SP100 proteins and the promotion of HCMV infection, as mentioned above.
Exhilaratingly, DNA-PKcs accompanied by X-ray repair cross-complementing protein 6 (XRCC6), splicing factor, proline-and glutamine-rich (SFPQ), RNA-binding protein 14 (RBM 14), Matrin-3, and Non-POU domaincontaining octamer-binding protein (NONO) was found binding to circSP100 in our study. These proteins were involved in the activations of the cGAS-STING-IRF3 pathway against DNA virus infection (Morchikh et al. 2017). Therefore, it was speculated that circSP100 might serve as a sponge to DNA-PK, which would block the combination of DNA-PK and HCMV DNA, resulting in inhibition of innate immunity against HCMV infection (Supplementary Fig. S2). In conclusion, it was demonstrated in our study that HCMV productive infection influenced host circRNA transcriptions in HELFs. Also, circSP100 transcribes differently from SP100 mRNA and potentially binds to 257 proteins. Nevertheless, the combination between circSP100 and the purified proteins needs to be proved, and the role of circRNAs during HCMV infection requires further investigation.
-
This work was supported by the National Natural Science Foundation of China (81672028 and 81371788).
-
JD, YH and QR conceived and designed the experiments. JD, QW, JL, YL and ZL performed the experiments. YM and YQ analyzed the data. JD and YH wrote the manuscript and prepared the figures. YM and YQ provided helpful suggestions about the study. QR checked and finalized the manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: