-
Canine parvovirus (CPV) belongs to the genus Parvovirus of Parvoviridae. CPV is thought to have emerged from feline panleukopenia parvovirus (FPLV) or a closely-related virus (10). Two antigenic variants, CPV-2a and CPV-2b, are now distributed worldwide. A third variant, CPV-2c, was detected in Italy in 2000 and is now circulating in Italy, Portugal, Germany and Vietnam together with types 2a and 2b (3, 6, 14).
CPV2 has a linear single-stranded DNA genome of 5323nt in length with hairpin structures at both ends (18). This virus has two major open reading frames (ORFs); one encodes two nonstructural proteins (NS1 and NS2) and the other encodes the capsid proteins VP1 and VP2. VP1 and VP2 are localized within the same DNA sequence and are translated from different start sites. Translation of VP2 begins at the residue 165 of VP1 (16, 18). VP3 is a truncated form of VP2 with approximately the first 20 amino acid residues lysised by proteinase(15). The virus particle consists of 60 subunits with no more than 10 VP1, the remainder comprising VP2 and VP3 (22). Langeveld et al. mapped the antigen epitopes of CPV2 using a complete set of overlapping nonpeptides (740 nonpeptides) of the capsid proteins VP1 and VP2. They found 10 antigenic sites six of which localized on the surface of the virus: three in the first 21 amino acid residues, one in loop1 (located in residue 100) and two in loop3 (residue 300) (11).
CPV2 can infect dogs at different ages with high infectivity, especially to young puppies. The best way for prophylaxis was vaccine inoculation. Traditional vaccines consist of either attenuated or inactivated microorganisms delivered by injection. Despite the low-virulence of the strain used, attenuated vaccines can cause severe diseases in immunodeficient individuals, and inactivated vaccines can still harbor toxins that compromise their safety. These problems can be circumvented by subunit vaccines. Research on synthetic peptide vaccines has shown that amino acid residues 2-21 of CPV VP2 are preferable for inducing neutralizing antibody (5). However, production of recombinant vaccine proteins in conventional expression systems is expensive, requiring large-scale fermentation equipments and stringent purification processes. These fermentation processes for vaccine production may be a prohibitively expensive technology for developing countries or for application in animal health. A plant expression system is an attractive alternative to conventional production systems to obtain recombinant proteins in large quantities at low cost, and is also free of human or animal pathogens and toxins (12). Fusion proteins of CPV amino acid residues 2-22 of VP2 (2L21) with either GUS (9) or CTB (13) were expressed in tobacco, respectively. Neutralizing antibodies were induced when animals were inoculated with these chimeric antigens (9, 13).
In this study, a strain of CPV2a was isolated and identified. Also, the N-terminal 414 amino acid residues of the vp2 gene that included the whole 6 exposed epitopes of CPV2 was cloned into a binary vector and used for tobacco transformation. Transgenic tobacco plants were obtained after selection on kanamycin-containing medium. The integration and expression of vp2 gene in transgenic tobacco was confirmed by PCR and Western blotting.
HTML
-
Materials and sources: DMEM medium (Sigma), Newborn calf serum (Gibcol, Invitrogen), trypsin (Difco). pBI121 vector (Clontech). Pre-stained protein ladder (SM0671, Fermentas). DNA Marker III and D15000+2000 (Tiangen Biotech Co., Ltd, Beijing). CPV Ag test strip (Anigen, Animal Genetics, Inc.). Madin-Darby canine kidney cell line was provided by Prof. Tianxian Li of Wuhan Institute of Virology, CAS. Feces samples of puppies were provided by Dr. Jianguo Chen.
-
Samples were collected from feces of sick puppies using cotton pledgets. Samples were washed into 1 mL phosphate buffered saline (PBS) in an 1.5 mL Eppendorf tube and centrifuged at 10, 000g for 5 min at 4℃. Supernatants were used to test virus titers by the hemagglutination test (HA) (4). One positive sample was filtered though a 0.22 μm membrane and 100 μL filtrate was added into a bottle of newly passaged MDCK cells and cultured with serum-free DMEM medium. CPV2 titer was detected every 24 h. Culture supernatants and MDCK cells were collected and lysed by freeze-thawing twice. CPV2 was collected from the supernatant by ultra-centrifugation at 200 000 g for 2 h at 4℃ after removing cell debris by centrifugation and with a 0.22 μm filter membrane.
-
The isolated CPV2 was identified with a commercial CPV Ag test strip, electron microscope and sequencing of its vp2 gene. The CPV Ag test was done according to the manufacturer's instructions. For more direct investigation, unstained CPV2 was detected by transmission electron microscopy. The vp2 gene was cloned by PCR using boiled viruses as template (7). Touch-down PCR was used with the following program: 94℃, 2.5 min; (94℃, 30s;60℃to 50℃, 30s, with temperature turn down 0.5 ℃ every cycle; 72℃, 1.5 min)×20;(94℃, 30s;50℃, 30s;72℃, 1.5 min)×10;and with a final extension at 72℃ for 7 min. The following primers were used: forward primer: 5'-atgagtgatggagcagttc-3', reverse primer: 5'-ttaatataattttctaggtgc-3'. PCR product was purified and then cloned into pMD-19-T (Takara) for sequencing (Invitrogen, Shanghai).
-
Four 6-8 weeks female BALB/c mice were immunized subcutaneouly at the nape and back by CPV2a emulsified with Freund's complete adjuvant. Two boosters were performed by intraperitonatal inoculation using CPV2a emulsified with Freund's incomplete adjuvant at two-week intervals. Ten days after the last booster, blood samples were collected from the orbital sinus and anti-CPV2 serum was obtained.
-
The coding sequence of the N-terminal 414 amino acid residues of vp2 was obtained by PCR with forward primer: 5'-TTTGGATCCATGAGTGATGG AGCAGTTC-3', and reverse primer: 5'-TTTGAGCTCCCAATCTCCTTCTGGATATCTTC-3'. A single 1 260 bp PCR product was purified and digested with BamH I and Sac I simultaneously. The digested fragment was inserted into pBI121 vector digested with the same restriction enzymes to construct plant expression vector pBI121-VP2. The resulting pBI121-VP2 was transformed into E. coli DH5α and selected on a Luria-Bertani (LB) plate with 100 μg/mL kanamycin. Plasmid extracted from E. coli DH5α was transformed into Agrobacterium tumefaciens strain EHA105 by electroporation.
-
Tobacco (Nicotiana tabacum cv. Xanthi) transformation followed the standard protocol (10). Primary transgenic plants were selected on MS medium containing 100 μg/mL kanamycin and 100 μg/mL timentin. Kanamycin-resistant plants were planted in the greenhouse. T1 transgenic tobacco plants with segregation against kanamycin were grown at 25℃ with 16 h light periods in a growth chamber and used for PCR analysis.
-
Western blotting of T0 transgenic lines. Total soluble protein (TSP) was extracted from tobacco leaf according to the method of Brodzik et al. (2) with the following modifications. Mature tobacco leaf was ground with 2 vol (v/w) of PBS buffer containing 0.5% Tween-20 (PBST) and 1mmol/L phenylmethyl sulfonylfluoride (PMSF). The mixture was centrifuged at 15 000g for 10 min at 4℃ and the supernatant was transferred to a new 1.5 mL Eppendorf tube. The supernatant was mixed with equal volume of 2×SDS-PAGE loading buffer and bathed for 5 min in boiling water. 10 μL sample was separated on SDS-PAGE gel and transferred to nitrocellulose (NC) membrane. After blocking in PBST buffer with 5% skimmed milk powder for 2 h, 200 fold diluted anti-CPV2 mouse serum in PBST was added and incubated at room temperature with continuous shaking for 1 h. 500 fold diluted AP-labeled goat anti mouse IgG (Vector) in PBST was added and incubated as above. After 4 times washing with PBST, NBT/BCIP substrate was added to develop color.
PCR detection of T1 transgenic tobacco. Genomic DNA of tobacco was extracted by the SDS method (21, 1) with the following modifications. 0.1 g of young tobacco leaf was ground into a fine powder after freezing in liquid nitrogen and transferred to a 1.5 mL Eppendorf tube. 0.5 mL extraction buffer (100 mmol/L Tris-Cl, 500 mmol/L NaCl, 50 mmol/L/L EDTA, pH 8.0 and 10% β-mercaptoethanol which is added before use) was added and vortexed for 10 s. 0.1 mL 10% SDS was added and mixed gently. 0.2 mL 5 mol/L KAC was added after bathing in 65℃ water for 20 min. After bathing on ice for 30 min, the supernatant was extracted once with an equal volume phenol/ chloroform/ isoamyl alcohol (25:24:1) and once with chloroform/ isoamyl alcohol (24:1). The aqueous phase was precipitated, washed with ethanol, dissolved in 30 μL of TE buffer and used for PCR analysis as described above.
Materials
Isolation and culture of CPV2
Identification of CPV2
Production of anti-CPV2 serum
Construction of vp2 expression vector for tobacco transformation
Tobacco transformation
Confirmation of vp2 transgenic tobacco plants
-
Four samples were collected from an animal hospital in Wuhan, China. One sample with a HA titer of 1: 20×27 was used for further analysis. Virus replication was detected by the HA test (Fig. 1A), CPV Ag strip assay (Fig. 1B), PCR (data not shown) and sequence analysis of the vp2 gene. This isolate was designated as CPV/WH02/06, and the GenBank accession number of its vp2 gene is EU377537. The amino acid residue 426 of VP2, which located on the surface of CPV2 capsid, can determine its neutralization characterization (26). According to the amino acid residue 426 of VP2, CPV2 is divided into 2a, 2b and 2c displaying Asn, Asp, and Glu, respectively (6). Residue 426 of VP2 of CPV/WH02/06 is Asn, with 99% homology to CPV-15 (GenBank accession M24003), which belongs to serotype 2a (16). We also made amino acid sequence alignment among VP2 CPV/WH02/06, CPV-15(2a), CPV V217 (2b, Accession No. AB054220) and CPV Uy-12/06 (2c, partial cds. Accession no. EF375479). Megalign of DNAstar 5.01 was used for sequence analysis. The amino acid residue 375 of VP2 can be used to differentiate CPV2 to CPV2 variants (2a, 2b, and 2c) with Asn and Asp respectively (26). The results of alignment confirmed that CPV/WH02/06 belongs to CPV 2a (Fig. 2). Condensed virus particles were also observed by transmission electron microscope with a diameter of ca. 20nm (Fig. 1C).
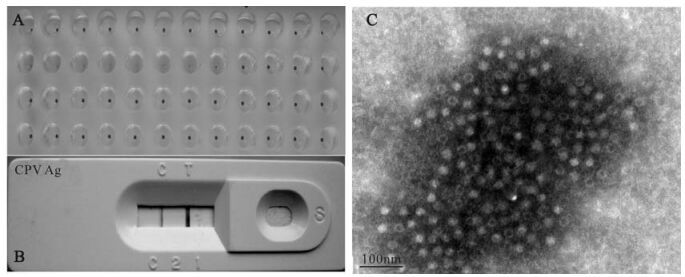
Figure 1. Identification of CPV2. A: Detection of CPV from feces samples of 4 dogs by HA test. B: CPV detection from culture supernatant of MDCK with dog feces sample by CPV Ag strip. C: Unstained CPV under transmission electron microscope.

Figure 2. Amino acid residue alignment of CPV2 by DNAstar 5.01 Megalign software. The amino acid residue 375 (shadow) of VP2 are all Asp (D) except CPV Uy-12/06, which only have partial coding sequence. The amino acid residue 426 is Asn (N), Asn (N), Asp (D), and Glu (E), which corresponding to CPV WH02/06, CPV-15, CPV V217, and CPV Uy-12/06, respectively.
-
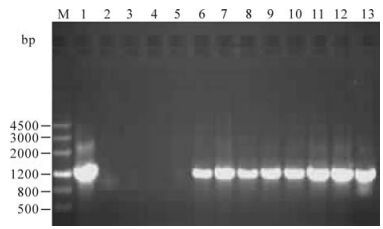
The plasmid of pBI121-VP2 was confirmed by restriction enzyme digestion analysis, and then immobilized into A. tumefaciens strain EHA105 for tobacco transformation. Independent kanamycinresistant plants were obtained and planted in the greenhouse to obtain T1 seeds. The seeds were germinated on kanamycin-containing medium to obtain the T1 plants. Four pBI121 empty vector transformed tobacco plants and 8 pBI121-vp2 independent transformed plants were tested for vp2 gene integration by PCR. As shown in Fig. 3, all of the 8 pBI121-VP2 transformed tobacco plants amplified a fragment of 1260bp size, which is the expected size of the vp2 gene.
-
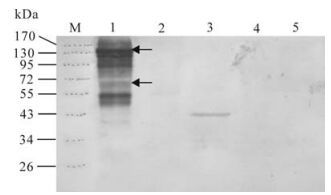
Three T0 transform tobacco lines were used to analyze VP2 expression by Western blotting using CPV2 virus as positive control and pBI121 transformed tobacco as negative control. As shown in Fig. 4, only one transgenic had the blot band of the truncated VP2, which corresponds to about 43 kDa. Line 1 contains blot bands of the CPV2 virus control, in which 130kDa, 65kDa is possibly corresponding to VP2 dimer and VP2 (25).
Isolation and Identification of CPV2
PCR detection of transgenic tobacco
Detection of VP2 protein in transgenic tobacco by Western blotting analysis
-
Although CPV2 has a DNA genome and uses cellular replication machinery, its rate of nucleotide substitution is closer to that of RNA viruses than to that of double-stranded DNA viruses (10). Since CPV2 was first found in 1978, three variants named CPV2a, CPV2b, CPV2c have been reported and replaced CPV2 as epidemic strains nowadays. In this report, we isolated a CPV2a serotype in Wuhan. This result suggests that CPV2a is still circulating in dogs in Wuhan. It is important to isolate epidemic virus strains and explore their potential for development of vaccines for control and treatment of diseases caused by this virus.
Wang et al. found that newborn calf serum (NCS) can inhibit CPV2 replication in MDCK cells and serum-free medium should be used in CPV2 culture (23). We obtained the same results and found that NCS has a high HA inhibition titer (1:50×24), so it is necessary to use serum-free medium for amplification of CPV2. There is no evidence of a cytopathic effect (CPE) in MDCK cells after infection with CPV2, so we monitored CPV2 replication by PCR and HA.
Treatment of CPV2 empty capsids can release oligomeric forms with the sizes of dimers, trimers, and pentamers of VP2 (25). In our experiment, most of the viruses were empty capsids with higher density core as shown in transmission electron microscope (Fig. 1 C). Virus sample was separated on SDS-PAGE without boiling in order to maintain the native antigen epitope, so the majority of the capsids were in oligomeric forms. The 130kd and 65kd blot bands are possibly the dimer or monomer form of VP2, respectively (Fig. 4). There are also many other blot bands on the CPV2 virus control line of the Western blot image. We consider that some of them are nonspecific reactions because the primary antibody was obtained by immunization of Balb/c mouse with the same virus sample.
In recent years, there has been considerable interest in the use of transgenic plants to generate compounds for medical and veterinary use. Advantages, as well as limitations, of using plants as recombinant protein expression system have been documented elsewhere (8, 24). Production of recombinant proteins in plants is economic, with minimal manufacture and processing requirements. Unlike mammalian based production systems, plant-derived antigens are free from the risk of contamination with animal pathogens. However, a major limitation of the use of transgenic plants for production of pharmaceutical proteins has been the relatively low level of expression (19). Transgene expression is influenced by several factors that cannot be controlled precisely through construct design, which leads to variable transgene expression and, in some cases, complete inactivation (17). Such factors include the positions of transgene integration, the structure of the transgenic locus, gene copy number and the presence of truncated or rearranged transgene copies. In our study, only one of the three PCR-positive transgenic lines was confirmed to have VP2 protein expression by Western blotting, this may relate to the low expression of the vp2 transgene or due to transgene silencing. Further studies are needed to obtain more transgenic lines and to test the immune responses of the expressed proteins in an animal model.














 DownLoad:
DownLoad: