-
Avian influenza virus (AIV) belongs to the Orthomyxoviridae family of RNA viruses. It is an enveloped virus with a helical nucleocapsid and eight segments of single-stranded negative-sense RNA. The envelope contains the haemagglutinin (HA) and neuraminidase (NA) proteins, of which there are currently 16 HA (H1-H16) and 9 NA (N1-N9) subtypes (1, 16). The H9N2 subtype that has spread worldwide in poultry can infect humans and is prevalent in China (2, 5, 6, 7, 12, 13). The first outbreak of disease attributed to H9N2 in China happened in Guangdong province in 1994. Other strains of this subtype were subsequently isolated (3, 7, 9, 12).
Studies to characterize these strains were focused on the HA genes for one of the surface viral proteins that plays a crucial role in the pathogenicity (9). Recent studies characterized the nonstructural (10) and neuraminidase genes (15) of H9N2 viruses in China. Others showed that some novel re-assorted H9N2 influenza viruses may possess certain genes from H5 subtype viruses (4, 8, 12). In this study, we have characterized three strains of H9N2 influenza viruses that were isolated from chicken farms in Guangxi province. All eight full-length genes of these isolates were obtained individually by means of RT-PCR. Sequence analysis and phylogenetic study were conducted by comparing the eight genes of each isolate with sequences available in Genbank (H9N2 isolates from 1966-2005).
HTML
-
Three isolates of H9N2 subtype influenza viruses, A/Chicken/ Guangxi/1/00 (C/GX/1/00), A/Chicken/ Guangxi/14/00 (C/GX/14/00) and A/Chicken/Guang-xi/17/00 (C/GX/17/00) were recovered from chickens in different areas of Guangxi province during 2000 and 2001. Initial isolation was performed in 10-day-old specific-pathogen-free (SPF) embryonated chicken eggs. Identification of the viruses was determined by standard hemagglutination-inhibition and neuraminidase-inhi-bition tests (17). Allantoic fluids were harvested from SPF egg-passaged viruses and used as stock viruses for further analysis.
-
Viral RNA was obtained from allantoic fluid by TRIzol extraction according to the method described by Xie et al. (18). The concentrations of RNA were determined by spectrophotometry using the UV2501PC (Shimadzu Cooperation, Tokyo, Japan) and then stored at −20 ℃. Reverse transcription was done by using random primers with RNA PCR (AMV) V3.1 kit (TaKaRa Biotechnology, Dalian, China). Amplifi-cation of the eight full-length genes was carried out by PCR as described previously by Xie et al (18) using pairs of specific primers as described in Table 1. The reaction was carried out at 30 ℃ for 5 minutes followed by 60 minutes incubation at 37 ℃. PCR was performed in a reaction mixture of 50 μL containing master mix buffer with dNTPs, 50-time Advantage 2 polymerase (Clontech Mountain View, California, USA), 10 μM of each specific primer, and 200 ng cDNA template. The PCR condition for the amplification of Pb2, Pb1, and HA was 95 ℃ for 2 minutes denaturation, and 35 cycles of 95 ℃ for 30 seconds, 62 ℃ for 45 seconds (annealing), and 68 ℃ for 3 minutes, followed by 70 ℃ for 10 minutes final extension. The PCR condition for the amplification of NP, NA, M, and NS genes was the same as above, except that the annealing temperature was reduced to 60 ℃ for NP and NA genes and 58 ℃ for M and NS genes.

Table 1. The nucleotide sequences of the eight pairs of specific primers for H9N2 subtypes.
-
All PCR products were subjected to electro-phoresis in a 1% (w/v) agarose gel. DNA fragments of the expected length were extracted and purified with the DNA Glass-milk Rapid Purification Kit (MK001-2, BioDev, Beijing, China). The purified DNA fragments were cloned into pMD18-T easy vector according to the manufacturer's protocol (TaKaRa Biotechnology Co. Ltd., Dalian, China). Three clones of each of the 8 fragments were sequenced by the Takara Biotechnology Co. Ltd., Dalian, China. DNA sequences of eight genes were compared with the GenBank database. DNA sequences of each cloned gene were repeated twice to confirm the similarity of sequence data. The nucleotide sequences obtained in this study are available in the GenBank database under accession numbers: DQ485205 to DQ485228.
-
The DNA sequences of the ORF nucleotide sequences of NA gene of three Guangxi H9N2 subtypes were compared initially with the Megalign program in the DNASTAR package using the Clustal alignment algorithm (DNASTAR Inc., Madison, Wis, USA) against the twelve H9N2 virus sequences as listed in Table 3. Pairwise sequence alignments were also performed with the Clustal alignment algorithm in the Megalign program to determine sequence similarity and the phylogenetic relationship of the different H9N2 subtype viruses. The ancestral relationships within eight genes among the seventeen H9N2 subtypes were presented in a phylogenetic tree created by the Megalign program.

Table 3. Comparison of ORF nucleotide sequences of NA gene; nt = nucleotide.
Viruses
RNA extraction and RT-PCR
Sequence Analysis
Phylogenetic Analysis
-
The lengths of all genes were obtained for the three isolates (C/GX/1/00, C/GX/14/00, and C/GX/ 17/00): HA 1.7 kb, NA 1.4 kb, NP 1.5 kb, M 1.0 kb, NS 890 bp, PA 2.2 kb, Pb1 2.3 kb, Pb2 2.3 kb. BLAST software and Megalign programs were used to determine the sequence similarity of the eight genes from the three isolates, as shown in Table 2.

Table 2. Length of each gene of AIV strains and Virus with the greatest homology to each gene
-
In this study, the nucleotide sequences obtained from these three H9N2 isolates were compared with those of all the available full-length genomes of H9N2 strains isolated during 1966-2005 and deposited in the GenBank database. Results indicate that nu-cleotide homology among these isolates with HA genes is between 82.6% to 99.3%; for those with NA genes the similarity is between 91.8% to 97.7%; for NP genes is between 88.0% to 98.7%; for M genes is between 92.7% to 99.6%: for NS genes is between 92.1% to 98.9%; for PA genes is between 86.1% to 94.1%; for Pb1 genes is between 86.8% to 96.2%; and for Pb2 genes is between 84.3% to 97.9%.
This sequence comparison also shows that 9 nucleotides between nucleotide positions 205 and 214, lying between position 187 to 195 open reading (ORF) in the NA genes, are absent in these three Guangxi isolates (Table 3) analyzed here. These 9 nucleotides encode for amino acids T, E, I. The deduced amino acid sequences of HA genes of C/GX/1/00, C/GX/14/00, and C/GX/17/00 at the cleavage site of HA contain a PARSSR/GL motif, which denotes the sequence found in low pathogenicity avian influenza (LPAI) viruses, as described previously (5, 9, 14).
-
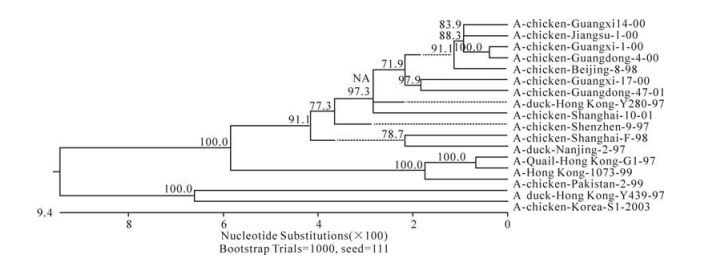
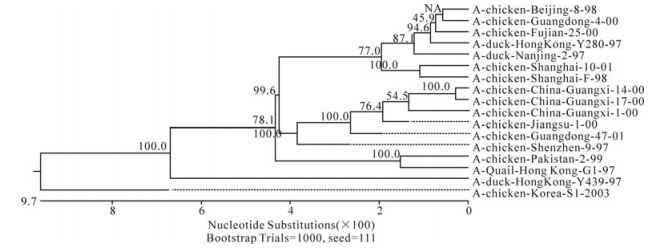
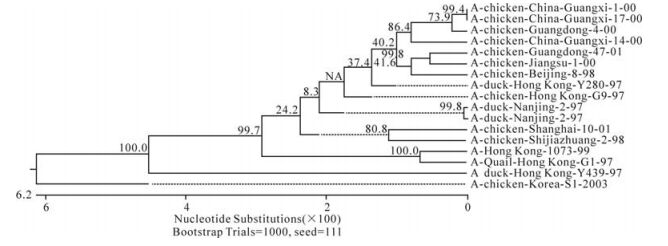
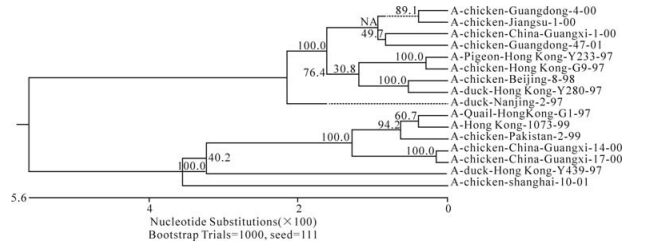
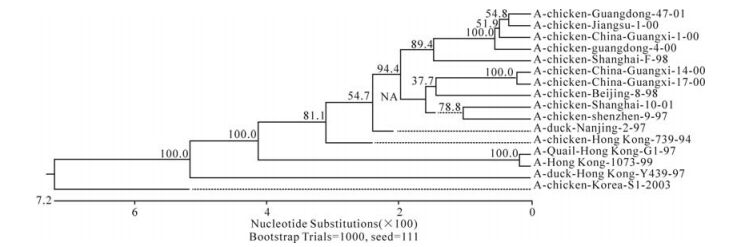
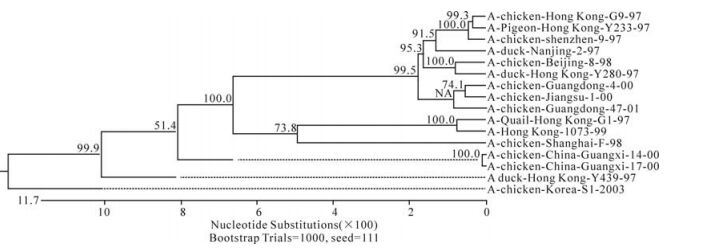
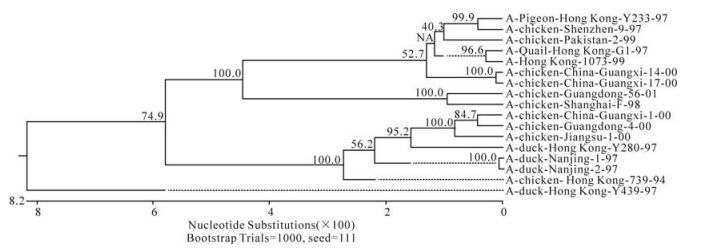
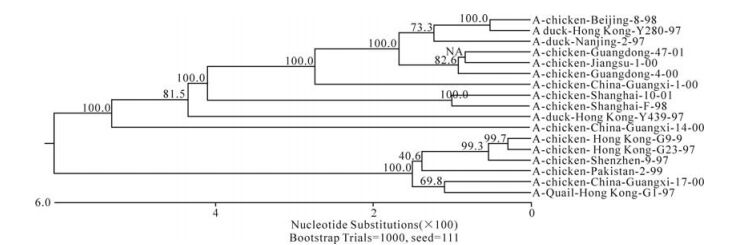
The phylogenetic relationship between each of the eight genes from three isolates, C/GX/1/00, C/GX/ 14/00, and C/ GX/17/00, was analyzed based on their nucleotide sequence (Fig. 1-Fig. 8). The analysis in-dicates that there are three sublineages in the Eurasian lineage as described previously (4, 10, 18). The NP and Pb1 genes of C/GX/14/00 and NP, Pb1, Pb2 genes of C/GX/17/00 were incorporated into the sublineage represented by A/Quail/Hongkong/G1/ 97 (Fig. 4, Fig. 7, and Fig. 8). The HA, M, NP, Pb1, Pb2 genes of C/GX/1/00, the HA and M genes of C/GX/14/00, and HA and M genes of C/GX/17/00 were incorporated into the sublineage represented by A/duck/Hongkong/Y280/97 (Fig. 1-Fig. 3). No gene of the three isolates was incorporated into the sublineage represented by A/duck/Hongkong/Y439/ 97. The PA, NA, NS genes of C/GX/1/00, NA, PA, NS, Pb2 genes of C/GX/14/00, and the NA, NS, PA genes of C/GX/17/00 were not incorporated into any of sublineage of these three isolates (Fig. 4, Fig. 5, Fig. 6, and Fig. 8).
Result of sequencing
Comparison of Nucleotide and Deduced Amino Acid Sequences of Eight Genes
Phylogenetic Analysis of Eight Genes
-
AIV has a segmented negative-strand genome that includes HA, NA, M, NS, NP, PA, Pb1, Pb2 genes. The results of sequencing and comparisons with the other H9N2 virus sequences showed that we have obtained each of eight genes from three Chinese isolates, C/GX/1/00, C/GX/14/00, and C/GX/17/00 successfully.
Previous studies have defined two distinct lineages of H9N2 influenza viruses: North American and Eurasian lineages. The Eurasian lineage consists of at least three sublineages (4, 8, 12). Phylogenetic analysis in our study showed similar patterns (Fig. 1-Fig. 8). Viral genomes of H9N2 viruses analyzed in previous studies had shown a common source of origin from southern China (3, 7, 9, 11). Sequence comparison and phylogenetic analysis illustrated that NP and Pb1 genes of C/GX/14/00, and NP, Pb1, and Pb2 genes of C/GX/17/00 were incorporated into the sublineage represented by A/Quail/Hongkong/G1/97. HA, M, NP, Pb1, and Pb2 genes of C/GX/1/00, HA and M genes of C/ GX/14/00, and HA and M genes of C/GX/17/00 were incorporated into the sublineage represented by A/duck/Hongkong/Y280/97. None of the genes from these three isolates was incorporated into the sublineage represented by A/duck/Hongkong/Y439/ 97. PA, NA, and NS genes of C/GX/1/00, NA, PA, NS, and Pb2 genes of C/GX/14/00, and the NA, NS, and PA genes of C/GX/17/00 were not incorporated into any of the sublineages. So, it is possible that AIV H9N2 subtype strains C/GX/1/00, C/GX/14/00, and C/GX/ 17/00 were products of natural reassort-ment of avian influenza viruses, suggesting that there may be a specific gene pool in China, as described by Lu et al (11).
Interestingly, gene sequence comparison indicates that there is a 9-nucleotide deletion between nuc-leotides 205 and 214 (187 position to 195 position of ORF) in the NA genes of the three isolates C/GX/1/ 00, C/GX/14/00, and C/GX/17/00. These 9 nucleotides encode for amino acids T, E, and I. This deletion did not change the ORF of the NA gene. Noteworthy, about 80% of China mainland isolates carry a similar deletion. Further studies are needed to determine if the lack of 9 nucleotides in NA affects the function of the NA protein.
The HA protein determines the infectious host range of AIV strains. AIV strains infecting humans carry leucine at position 226 of the HA gene (4, 8, 12). The analysis of deduced amino acid sequence of HA proteins of C/GX/1/00, C/GX/14/00, and C/GX/ 17/00 isolates showed a glycine at position 226, therefore these three isolates do not belong to H9N2 AIV strains isolated from human infections.














 DownLoad:
DownLoad: