-
The complex and singular life cycle of polydnaviruses (PDVs) has fascinated biologists ever since these unusual viral entities were first reported in the scientific literature. As such, they have raised countless questions, many of which have been addressed through experimental work focusing on the elucidation of their functions and origins.
PDVs are dsDNA viruses whose genome is made up of multiple circular segments. Their replication is confined to the ovaries of some endoparasitic wasps, where viral DNA is generated from a copy of the viral genome permanently maintained within the wasp genome. Virions are assembled in the nuclei of ovarian calyx cells and subsequently released into the lumen of the oviducts. They are later injected into a lepidopteran host during the process of parasitization (i.e., egg laying); in this host, no viral replication takes place but expression of PDV genes induces immune and developmental disturbances that are essential to the successful completion of wasp development. For this reason, the association of PDVs with parasitic wasps has been described as mutualistic (14, 22).
Recent endeavors in the area of PDV genome sequencing and annotation (7, 12, 16, 23, 26) have generated a wealth of data and new hypotheses about the evolution of these intriguing insect viruses, as well as new questions about the diversification and functions of the new putative genes identified in their genomes.
In the three campoplegine ichnoviruses (IV) (PDVs associated with ichneumonid wasps of the subfamily Campopleginae) whose genomes have been sequenced [Campoletis sonorensis IV (CsIV), Hyposoter fugitivus IV (HfIV) and Tranosema rostrale IV (TrIV)] (23, 26), approximately half of the predicted ORFs have been assigned to previously described or characterized gene families, such as those encoding proteins that display significant sequence or structural similarity to proteins found in other organisms (inx, ank, and Cys-motif), while members of the remaining families were identified on the basis of similarity to previously characterized IV transcripts (rep and TrV families) or because of the demonstrated existence of related putative ORFs among two or more IV genomes (N, PRRP). All other putative ORFs, which constitute the remaining half, could not be readily assigned to specific gene families because they did not display similarity to known proteins ("unassigned" ORFs).
Annotation of the TrIV genome revealed the presence of several gene families. The repeat element family (rep) is the largest with 17 members, followed by the TrV family (7 members), N family (4 members), inx family (3 members), ank family (2 members), Cys-motif (1 member) and PRRP (1 member). The remaining putative ORFs (59%) could not be assigned to any known family (23).
In earlier studies, we assessed the transcription of selected TrIV genes from the rep (TrFrep1) (25) and TrV (TrV1, TrV2 and TrV4) (1, 2, 5) families in the lepidopteran host Choristoneura fumiferana by Northern blot analysis. More recently, we conducted a detailed qPCR analysis of the abundance of all 17 TrIV rep transcripts, in both lepidopteran and wasp hosts (19). This study indicated that two TrIV rep genes, F1-1 and F1-2 (= TrFrep1 and TrFrep2), are expressed at much higher levels than all other members of this family in infected C. fumiferana larvae. In addition, the rep transcriptional profile seen in T. rostrale ovaries was found to be markedly different from that observed in infected caterpillars.
For the present study, we wanted to extend the latter qPCR analysis to other putative ORFs identified during annotation of the TrIV genome, so as to assess the accuracy of our gene predictions and to generate a global transcriptional profile for a large sample of TrIV genes across all known families and among unassigned genes. Here, we show that a high proportion of genes identified during annotation are expressed in either the caterpillar or wasp (ovaries) host, but that some members of the TrV and rep families are expressed at much higher levels in infected caterpillars than genes from any other TrIV gene family examined, suggesting that selected members of these two families play a critical role in host subjugation. Similarly, the transcripts generated by another rep gene and a previously unassigned gene clearly outnumber all other TrIV transcripts in wasp ovaries. This previously unassigned gene is shown to belong to a new family of four genes encoding secreted proteins expressed almost exclusively in wasp ovaries and displaying a novel cysteine motif.
HTML
-
Choristoneura fumiferana larvae were either parasitized by T. rostrale within 24 h after the molt to the last instar or injected with 0.5 female equivalents (FE) of calyx fluid (CF), as described (9, 10). Total RNA was extracted from five larvae of each group 3 d post-parasitization (p.p.) or post-injection (p.i.), using TRIZOL reagent (Invitrogen), according to the manufacturer's instructions (1). In addition, total RNA was extracted and pooled from five ovary pairs dissected from post-emergence 5-10 day-old T. rostrale females, using the QIAshredder and RNeasy Mini Kit (Qiagen), according to the manufacturer's instructions.
-
A cDNA library was constructed as described (18) using RNA extracted from CF-injected C. fumiferana larvae. Briefly, 3 µg of total RNA was reverse-transcribed using an oligo-dT primer with the following sequence: TTTTGTACAAGC (T)16, followed by synthesis of the second cDNA strand and ligation of an adaptor; the latter was used for amplification of the cDNA using an adaptor-specific primer (ASP; 5x-CTAATACGACTCACTATAGGGC-3x) in conjunction with the oligo dT primer. PCR amplification was performed using 0.1 µmol/L of primers, 0.3 mmol/L of each dNTP and 1.5 U of Taq platinum High Fidelity (Invitrogen) in 1×PCR High Fidelity buffer (Invitrogen), containing MgSO4 (2 mmol/L). The conditions consisted of a first heating step at 94℃ for 2 min, and then 20 cycles of 94℃, 30 s; 55℃, 1 min; 68℃, 5 min.
-
To determine whether some of the TrIV ORFs that had not been assigned to a known gene family (23) could form new families, we conducted local blast (Blastp) searches against a TrIV unassigned ORF data base, followed by a multiple amino acid sequence alignment performed by ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html), subsequently adjusted manually for one of the identified families. For amino acid composition analysis and signal peptide predictions, we used ProtParam (http://www.expasy.ch/tools/protparam.html) and SignalP (http://www.cbs.dtu.dk/services/SignalP/), respectively. Disulfide bond predictions were made using the Scratch Protein Predictor (http://www.ics.uci.edu/~baldig/scratch/).
-
To determine which of the putative ORFs identified in the genome of TrIV were expressed in TrIV-infected larvae, we first conducted PCR amplifications of predicted TrIV ORFs from the above-mentioned cDNA library. Primers were designed within the coding sequence of each putative ORF (Table 1). 2 µL of a 25× dilution of the cDNA library was used for PCR amplification, with 0.25 µmol/L of each primer and 0.2 mmol/L of each dNTP, in 1× PCR buffer. After a hot start at 94℃ for 3 min, PCR was carried out by addition of 2 U of Tag DNA polymerase at 80℃. The rest of the cycling conditions were as follows: 30 cycles of 94℃, 45 s; 48℃, 45 s; 72℃, 1 min; and a final extension step at 72℃ 5 min. The amplification products were then cloned into pGEM-T easy vector (Promega) according to the manufacturer's instructions and sequenced.

Table 1. Oligonucleotide sequence and orientation of primers designed for PCR amplification of TrIV putative ORFs from a cDNA library
-
To remove DNA contaminants from RNA extracts, 500 ng of total RNA was treated with 2 U amplification-grade DNase I (Invitrogen) for 15 min at 25℃. We ran no-RT controls for the four most highly transcribed ORFs and detected no significant amplification, pointing to the virtual absence of genomic DNA contamination in the extracts. RNA (500 ng) from parasitized and CF-injected C. fumiferana larvae, and 200 ng RNA from ovarian tissue were reverse-transcribed using 0.5 µg of an oligo(dT) primer and 200 U Superscript II RNase H- reverse transcriptase (Invitrogen). The reaction was carried out in 1×PCR buffer, with 0.5 mmol/L of each dNTP and 40 U of RNAguard ribonuclease inhibitor (Amersham Biosciences), at 42˚C for 50 min.
For qPCR analysis, four primers were initially designed for each TrIV gene, using diverse regions among aligned nucleotide sequences. These four primer pairs were used to assess primer performance and quantitative precision. Initial amplification tests were conducted on reverse-transcribed RNA obtained from parasitized C. fumiferana larvae. A single primer pair was then selected for each gene (Table 2), based upon high amplification efficiency and lack of non-specific amplification products, and used for the analysis of the remaining samples.

Table 2. Oligonucleotide sequence and orientation of primers designed for quantitative real-time RT-PCR (qPCR) amplification of TrIV putative ORFs.
PCR amplifications were carried out on aliquots of individual RT reactions containing cDNA in amounts equivalent to 2.5 ng RNA, except for ovarian samples, which contained amounts of cDNA equivalent to 1 ng RNA. Four replicate amplification reactions containing 500 nmol/L of each primer were conducted for each sample, using an MX3000P spectrofluorometric thermal cycler (Stratagene) and QuantiTectTM SYBR Green PCR Kit (Qiagen), initiated with a 15-min incubation at 95℃, followed by a cycling regime of 95℃, 10 s and 65℃, 2 min. Each run was completed with a melting curve analysis to confirm the specificity of amplification and absence of primer dimers. Amplification efficiency was determined for each amplification reaction using LRE ("linear regression of efficiency") analysis, and the number of target molecules calculated using lambda genomic DNA as a quantitative standard (19-21).
RNA extraction
cDNA library construction
Bioinformatics analyses
Amplification of ORF-specific cDNAs from the cDNA library
Reverse transcription and quantitative real-time PCR (qPCR)
-
As a first step towards determining which of the known and putative TrIV genes are expressed in infected C. fumiferana hosts, we conducted ORF-specific PCR amplifications from a cDNA library constructed using RNA from TrIV-injected C. fumi-ferana last instars, 3 d p.i. Using this approach, transcripts were detected for 77% of all assigned TrIV ORFs, while only 42% for the 12 unassigned ORFs that we sampled generated amplification products (Table 3). These proportions increased to 91% and 67%, respectively, when the presence of gene-specific transcripts was assessed using the more sensitive qPCR-LRE approach (Table 3). Thus, the vast majority of TrIV genes assigned to specific families during genome annotation were found to be expressed in TrIV-infected C. fumiferana larvae; for unassigned genes, this proportion was lower, based on the present sample. Furthermore, as indicated in the quantitative analyses presented below, some TrIV genes were found to be expressed almost exclusively in T. rostrale ovaries.

Table 3. Overall assessment of the expression (detected or not; + or -) of known and predicted TrIV ORFs in TrIV-infected C. fumiferana larvae.
-
Although none of the 11 TrIV genes identified as belonging to the ank, inx, Cys-motif, PRRP and N families displayed very high levels of transcripts in either infected C. fumiferana hosts or T. rostrale ovaries (≤3 000 transcripts/ng total RNA), six of them had more abundant transcripts in wasp ovaries than in parasitized caterpillars, including two ank, two inx and two N genes (Fig. 1). With the exception of the C6-1 and D4-1 inx genes, this inter-host difference was less pronounced when the comparison was made with transcript levels measured in virus-injected caterpillars, presumably as a result of the supraphysiological viral dose present in 0.5 FE of calyx fluid (19). Interestingly, the only member of the Cys- motif family identified in the TrIV genome was expressed at very low levels ( < 200 transcripts/ng total RNA) in both infected caterpillars and wasp ovaries, while transcript abundance for the single TrIV representative of the PRRP gene family (23) was moderate (~700-3000 transcripts/ng total RNA) in the three samples examined (Fig. 1).
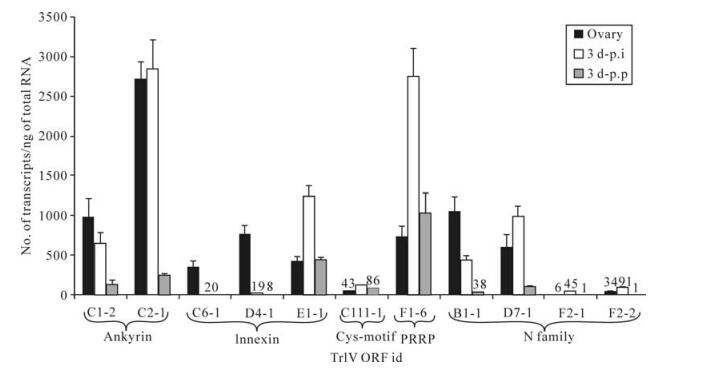
Figure 1. qPCR determination of transcript levels of 11 TrIV putative genes (23), distributed among five gene families, in C. fumiferana 6th instar larvae, 3 d following natural parasitization by T. rostrale (3 d p.p.) or injection of 0.5 FE T. rostrale calyx fluid (3 d p.i.), as well as in T. rostrale adult ovaries. Larvae were parasitized or injected within 24 h after the molt to the 6th instar. For each measurement, total RNA was extracted and pooled from 5 larvae or 5 ovary pairs dissected from 5-10 day-old females. Actual transcript numbers are provided above each bar for values < 100. Each value presented here is the mean of four technical replicates carried out on each RNA extract. Error bars: SD.
-
Prior to generating estimates of transcript abundance for a sample of genes among the 51 unassigned TrIV ORFs identified earlier (23), we wanted to determine whether some of these genes formed families. Given that PDV genes tend to fall within families of related coding regions, we reasoned that putative ORFs that had clear relatives within the TrIV genome were more likely than orphan ORFs to be real genes (i.e., transcribed DNA). Local Blastp analyses led to the identification of three small groups of related proteins encoded by unassigned ORFs (Fig. 2). The first of these groups contains four members, all of which display a novel C-terminal cysteine motif. Thus, to obtain a preliminary assessment of the transcriptional activity of TrIV unassigned genes, we measured transcript levels for six ORFs randomly selected among those that were considered orphans and for six others that appeared to belong to a gene family (i.e., those presented in Fig. 2A and B). Interestingly, five of the six orphan ORFs had barely detectable transcripts, whether in infected hosts or in wasp ovaries, while the remaining orphan gene had low but detectable quantities of transcripts in wasp ovaries (~500 copies/ng total RNA). In contrast, the four members of the family shown in Fig. 2A displayed moderate levels of transcripts (~2 000-12 000 copies/ng total RNA) in wasp ovaries, while being expressed at very low levels in infected caterpillars (Fig. 3). For this reason, these proteins are here assigned to a new TrIV gene family, designated "Ovary-Specific Secreted Proteins" (OSSPs). The other two related proteins examined were also expressed almost exclusively in wasp ovaries, but at levels lower than those measured for OSSPs.
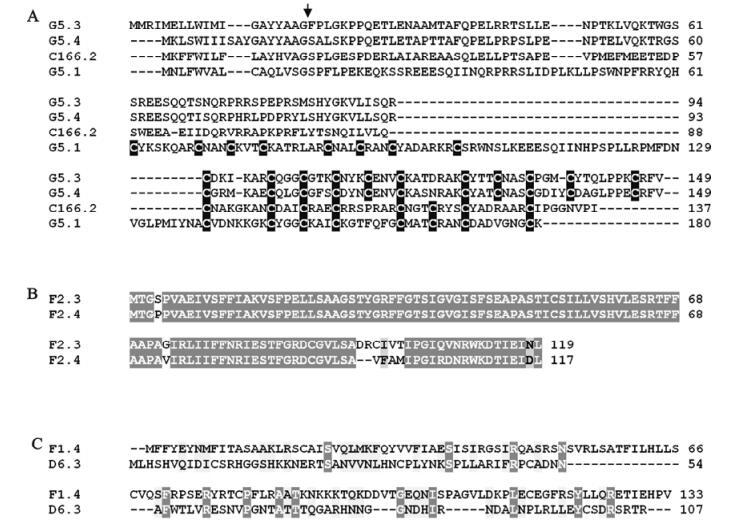
Figure 2. ClustalW alignment of amino acid sequences deduced from selected TrIV unassigned ORFs that were found to form groups of two or more related proteins. A : Four related proteins displaying a novel C-terminal cysteine motif (cysteine residues are shown as white letters against black background). The arrow indicates the position of the putative signal peptide cleavage site. B: Two very similar proteins encoded by unassigned ORFs found on genome segment F2. This group is here designated "unassigned family B". C: Two proteins encoded by unassigned ORFs and displaying modest similarity. For B) and C), identical residues are shown as white letters against dark gray background, while similar residues are shown as black letters against light gray background.
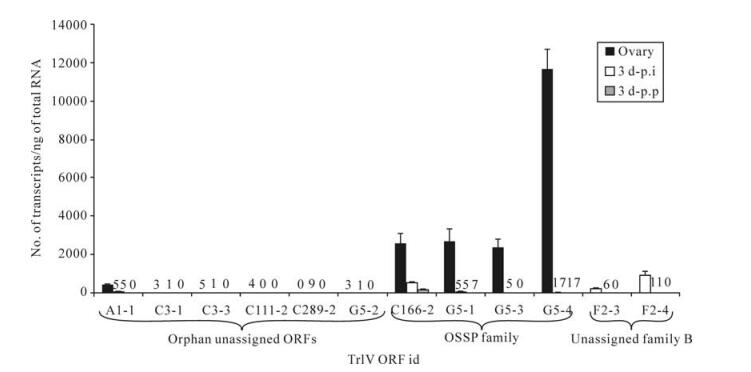
Figure 3. qPCR determination of transcript levels of 12 TrIV putative ORFs selected among 51 unassigned ORFs (23), in C. fumiferana 6th instar larvae, 3 d following natural parasitization by T. rostrale (3 d p.p.) or injection of 0.5 FE T. rostrale calyx fluid (3 d p.i.), as well as in T. rostrale adult ovaries. Putative genes are here clustered according to whether they are orphan or belong to a family ("OSSP" and "unassigned family B"; see caption of Fig. 2). For each measurement, total RNA was extracted and pooled from 5 larvae or 5 ovary pairs dissected from 5-10 day-old females. Larvae were parasitized or injected within 24 h after the molt to the 6th instar. Actual transcript numbers are provided above each bar for values < 100. Each value presented here is the mean of four technical replicates carried out on each RNA extract. Error bars: SD.
-
To estimate the relative importance of each gene family with respect to the abundance of their transcripts in infected caterpillars and wasp ovaries, we selected, for each family, the gene for which the highest level of transcripts had been measured in TrIV-injected C. fumiferana last-instar larvae, 3 d p.i., or in adult wasp ovaries (Fig. 4 and Fig. 5). In infected caterpillars, TrV1 (a detailed transcriptional analysis of genes from the TrV family will be reported elsewhere), which encodes a secreted protein, was by far the most highly transcribed TrIV gene, with nearly 300 000 copies/ng total RNA (Fig. 4). The rep family came second in this ranking, with the F1-1 gene (TrFrep1) producing ~52 000 transcripts/ng total RNA. In comparison, ank-2, PRRP and inx-3 generated transcript quantities varying between ~1 000 and 3 000 copies, while all others produced < 1 000 copies/ng total RNA (Fig. 4).
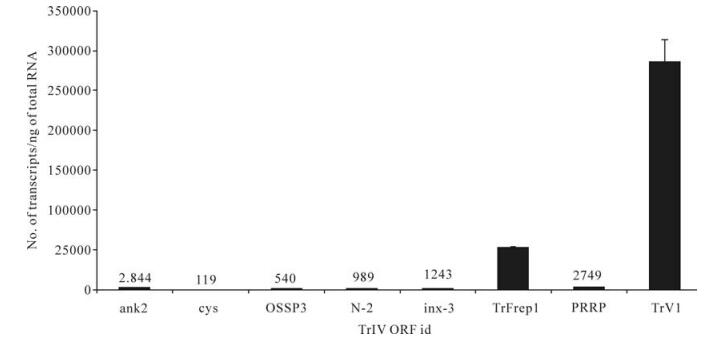
Figure 4. Comparison of transcript abundance among selected representatives of all known TrIV gene families, in C. fumiferana 6th instar larvae, 3 d following injection of 0.5 FE T. rostrale calyx fluid (3 d p.i.). Larvae were injected within 24 h after the molt to the 6th instar. For each family, we show the value obtained for the most highly transcribed gene in infected caterpillars. For each qPCR measurement, total RNA was extracted and pooled from 5 larvae. Actual transcript numbers are provided above each bar for values < 50, 000. Each value presented here is the mean of four technical replicates carried out on each RNA extract. Error bars: SD. Data for TrFrep1 are from Rasoolizadeh et al. (19).
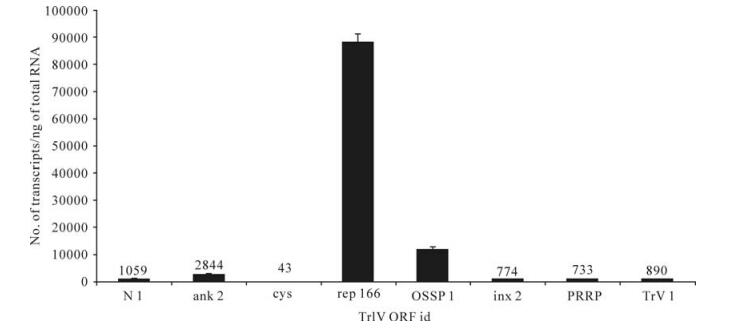
Figure 5. Comparison of transcript abundance among selected representatives of all known TrIV gene families, in adult T. rostrale ovaries. For each family, we show the value obtained for the most highly transcribed gene in this tissue. For each qPCR measurement, total RNA was extracted and pooled from 5 ovary pairs dissected from 5-10 day-old females. Actual transcript numbers are provided above each bar for values < 10 000. Each value presented here is the mean of four technical replicates carried out on each RNA extract. Error bars: SD. Data for rep166 are from Rasoolizadeh et al. (19).
In wasp ovaries, a rep gene (C166.1 or rep166) dominated the transcriptional profile, with ~90 000 copies/ng total RNA, followed by OSSP1, which had ~12 000 copies (Fig. 5). For all other genes, transcript abundance was≤1 000 copies/ng total RNA, except for ank-2, which generated ~2 800 copies (Fig. 5).
-
In the course of annotating the TrIV genome, seven genes were identified as being spliced (Cys-motif, TrV1, TrV2, TrV3, TrV4, TrV5 and TrV6), all of which are predicted to encode secreted proteins (23). The splicing junctions of three of these, TrV1, TrV2, and TrV4 had been confirmed in earlier studies (1, 2). Here, we attempted the cDNA cloning and sequencing of the remaining four genes to determine if they were indeed spliced and whether the splicing junctions had been predicted correctly. We were not able to amplify TrV5 and TrV6 from our cDNA library or by qPCR (Table 3), suggesting that these two very small putative ORFs (they encode proteins of 74 and 56 amino acid residues, respectively) may well be pseudogenes. However, we were able to clone the cDNAs of the Cys-motif and TrV3 genes, both of which were confirmed to contain two exons and one intron, although the length of the first exon had been incorrectly predicted in both cases (Table 4); corrections have now been made to the appropriate GenBank entries.

Table 4. Differences between predicted and observed splicing junctions for two TrIV spliced genes, TrV3 and a Cys-motif gene.
Detection of TrIV transcripts in infected larvae
Transcript abundance of TrIV ank, inx, Cys-motif, PRRP and N genes
Transcript abundance of TrIV "unassigned" genes
Comparison of transcript abundance across all TrIV gene families
Accuracy of splicing junction predictions
-
Transcriptional analysis often constitutes a first step towards identifying the function of a gene. The present study, along with an earlier one focusing on rep genes (19), provides a global assessment of transcript abundance, in both infected lepidopteran hosts and carrier wasp ovaries, for more than half of the genes identified in the genome of the ichnovirus TrIV (23). As such, this analysis makes it possible to evaluate the likely importance of genes within each of the known ichnovirus gene families, as may be inferred from their observed levels of expression.
The quantitative data presented in Fig. 4 confirm earlier assessments made by Northern analysis (1, 2, 5) to the effect that TrV1 is, by far, the most highly expressed TrIV gene in infected C. fumiferana larvae, with transcript levels almost six times higher than those of the most highly expressed rep gene, TrFrep1. In comparison, genes from all other families are expressed at levels > 15 times lower than those of TrFrep1 (Fig. 4). These results suggest that genes from the TrV and rep families, and more specifically TrV1 and TrFrep1, encode products that are likely to be required for induction of developmental arrest, which is the principal physiological perturbation observed in TrIV-infected C. fumiferana hosts (9, 10).The Cys-motif gene family (8) has ten representatives in the CsIV genome (26), some of which are abundantly expressed in parasitized Heliothis virescens larvae (3, 4). In this host, their protein products appear to play a role in both immune suppression (6, 17) and developmental disturbances (13). In comparison, we detected only one member of this family in the TrIVgenome (23), and its expression was here observed to be very low in the three samples we examined (Fig. 1). These results support our earlier hypothesis (23) that Cys-motif genes may no longer be required by T. rostrale to achieve successful parasitism, inasmuch as TrIV has little or no impact on the cellular immune response of C. fumiferana hosts (9, 10). However, it has been noted earlier that Cys-motif and TrV genes appear to have a common ancestor (1), but that TrV proteins lack the characteristic cysteine motif (C… C…CC…C…C) of CsIV Cys-motif gene products, which may be essential to achieve host immune dysfunction, while it may not be required for induction of developmental arrest.
The only other ichnovirus gene family that has been extensively examined with respect to transcript abundance is the ank (or vank) family in CsIV (15). Although transcript levels of the seven known CsIV ank genes were not compared with those of other CsIV genes, all but one were readily detectable by Northern blot analysis of RNAs extracted from parasitized H. virescens larvae, and two of the protein products could be detected by either Western blot analysis or immunofluorescence assays (15). In addition, using an rq-RT-PCR strategy, transcripts of all seven genes could be quantified in parasitized larvae, and transcripts could also be detected at low levels in female wasps (15). In the present study, the two known TrIV ank genes were expressed at higher levels in wasp ovaries than in parasitized C. fumiferana hosts at 3 d p.p. (Fig. 1), but transcript abundance was below 3, 000 copies/ng total RNA in both hosts. Of course, our sampling time may not have been optimal for the detection of TrIV ank transcripts in C. fumiferana, although CsIV ank mRNA levels were typically maximal at 3 d p.p. in parasitized H. virescens larvae (15). In addition, the higher transcript levels observed in female wasps, compared with parasitized caterpillars, may be due, at least in part, to the fact that we limited our analyses to wasp ovaries, thereby generating an RNA sample enriched in TrIV transcripts, as the ovaries appear to be the only tissue supporting significant TrIV gene transcription in the T. rostrale host (19). Nonetheless, the data presented here suggest that TrIV ank genes could play a limited role in altering C. fumiferana host physiology.
It has been known for many years that some ichnovirus genes are expressed in the reproductive tract of female wasp carriers (4, 24), although the functional significance of such expression has not been elucidated. As reported earlier (19). one of the 17 TrIV rep genes, rep166 (C166.1), was transcribed at relatively high levels in T. rostrale ovaries, while transcript abundance of TrIV genes associated with other ichnovirus families identified prior to the present study, including the TrV family, was much lower (Fig. 5). However, transcript levels of OSSP1, one of the four members of a novel TrIV gene family (Fig. 2), were sufficiently high (~12, 000 copies/ng total RNA) to make us consider the possible role of this protein in the biology of T. rostrale. Since OSSPs are predicted to be secreted, they could accumulate in the lumen of the oviduct prior to being injected into the lepidopteran host during oviposition. Their C-terminal cysteine motif is clearly distinct from that of ichnovirus Cys- motif proteins, but the disulfide bonds they are predicted to form should ensure their stability until injection into the lepidopteran host, in which they could play a role in host regulation before TrIV gene expression begins. Unlike OSSP1, rep166 is not a secreted protein, and is therefore not predicted to accumulate in the ovarian fluid. For this reason, we have suggested that it may play a role in virus replication (19). Hypotheses regarding the roles of these two proteins are currently being addressed experimentally.
In addition to generating a global profile of TrIV gene transcription in infected C. fumiferana larvae, the present study provides an assessment of gene predictions made during annotation of the TrIV genome (23). Overall, these predictions were accurate, particularly in the case of ORFs that could be assigned to known ichnovirus gene families (Table 3), although small errors were made in identifying the splicing junctions of two genes (Table 4). With respect to "unassigned" ORFs, our predictions appear to have been somewhat less accurate, particularly for "orphan" putative genes, although this conclusion is based on a relatively small sample of genes. It should also be pointed out that the few genes that escaped detection in the present study could well be expressed in other lepidopteran hosts of T. rostrale.














 DownLoad:
DownLoad: