-
Chilo iridescent virus (CIV) or Invertebrate irides-cent virus 6 (IIV-6) was originally isolated from diseased larvae of the rice stem borer, Chilo suppres-salis (Lepidoptera; Pyralidae) in Japan (29), and can infect many insect species. The term iridescent in the virus name is due to the turquoise colour of heavily infected larvae. The massive proliferation of CIV particles and their paracrystalline arrangement in the cytoplasm of host cells cause this turquoise color, which is indicative of a patent infection that is usually fatal. The colours include lavender and blue. However patent CIV infections are usually rare. Concentrated samples of purified virus from sublethally infected insects may also iridescence (62).
CIV has been most studied in the respect of host range and has been shown to cause patent infections in numerous species from the major insect orders, in-cluding species of agricultural and medical impor-tance (28, 52) and a number of arthropods (53).
The large DNA genome of CIV has been totally sequenced, and several genes have been characterized at transcriptional levels (39, 22, 23). Recently the molecular aspects of CIV have been the subject of a number of studies (16, 36, 49, 50, 56,).
HTML
-
The family Iridoviridae comprises icosahedral particles with an internal lipid membrane and a large double stranded DNA genome (14). There are cur-rently five genera recognized in this family (Table 1). Invertebrate iridescent virus 6 (IIV-6), often referred to as Chilo iridescent virus (CIV) is the type species of the Iridovirus genus which comprises one additional species: Invertebrate iridescent virus 1 (IIV-1).

Table 1. Taxonomy of iridoviruses
CIV has been used as the standard model for studies on invertebrate iridoviruses. Besides CIV and IIV-1, there are additional 11 tentative species recognized in the genus. However currently, there is not enough information available to determine their species status (63). Williams and Cory (64) noted that at least two strains of CIV being used in laboratories in different parts of the world. The isolate that has been com-pletely sequenced by Jakob et al. (39) in Germany differs from the isolates used in New Zealand, Australia, and the USA.
-
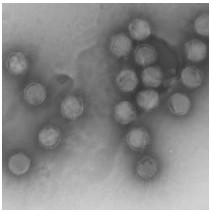
CIV is a large non occluded virus with icosahedral symmetry (Fig. 1). The virion is composed of three concentric domains; an outer proteinaceous capsid, an intermediate lipid membrane with associated poly-peptides, and a central core DNA-protein complex containing the genome (14, 62, 63). Viral particles can be either enveloped or nonenveloped, depending on whether they are released from the cell by lysis or bud from the plasma membrane.
CIV particle structure has been defined by cryo-electron microscopy and three-dimensional image reconstruction. Image reconstruction shows that the CIV capsid (185 nm maximum diameter) consists of 12 pentamers 1460 doughnut-like trimers (each trimer has a fiber that emanates from the center) arranged in a T=147 icosahedral lattice. A lipid bilayer follows the inner side of the capsid. Also the CIV capsid consists of 20 trisymetrons and 12 pentasymetrons and each trisymetrons contain 55 trimers (67). In a recent study the structure of CIV was determined to high resolution (13 Å ) by means of cryo-electron microscopy [cryo-EM] and three-dimensional image reconstruction. These studies revealed that there are at least three different types of minor capsid proteins associated with the capsomers outside the lipid membrane of CIV (68).
The structure of the internal lipid membrane has been investigated and noted to be synthesized de novo in the infected cell cytoplasm without any visible continuity with the cell membrane (58). Another group investigated the lipid composition of the CIV. They found that the lipid composition of the viral membrane was unchanged whether the virus was propagated in vivo in larvae or in vitro in invertebrate cell cultures and was clearly different from that of hosts. A high abundance of phosphatidylinositol and diglycerides was detected in compositional analyses, indicating that CIV preferentially incorporates these lipids into its internal membrane (1).
-
The host range of CIV in the class Insecta has been investigated by intrahemocoelic or per os inoculation and found to include more than 100 insect species belonging to six orders (Lepidoptera, Coleoptera, Diptera, Hymenoptera, Hemiptera and Orthoptera) (28, 32, 33, 47, 48, 52, 53). Fukuda (28) succeeded in perorally infecting 13 species of mosquitoes with CIV. Ohba (52) infected 65, 8 and 2 species of Lepidoptera, Coleoptera and Hymenoptera, respectively.
CIV replicates in many insect cell lines (13, 18, 41, 52) including cells from Bemisia tabaci (30), Diaprepes root weevil (35), and the boll weevil (24), and can even infect reptile cells (46). However, CIV does not replicate in vivo in frogs (54). Following the in-traperitoeal injection into Rana limnocharis, the titer of CIV declined 100 fold over a period of four days. In a recent study, the cell line AFKM-On-H, from hemocytes of the European corn borer Ostrinia nubilalis (Hübner) (Lepidoptera, Pyralidae) was also studies for its ability to productively sustain CIV infection (5). Several ultrathin sections demonstrated internalization of virus particles in cytoplasm of cells without clear signs of CIV replication. However, suspicions were raised concerning the toxicity of the virus because cells were inoculated with high con-centrations of CIV and since CIV readily infects O. nubilalis and replicates abundantly in different tissues and cells of larvae including the fat tissue and hemocytes (4).
In spite of its wide host range CIV has attracted little attention as a potential biopesticide, because of the limited mortality effect on its hosts (63). However, non-lethal inapparent infections may be common in certain insect populations (60, 61) and such infections may seriously reduce the reproductive capacity, body size and longevity of infected individuals. Little is known about the factors that determine the virulence of these viruses, but it is assumed that covert infections open the way to vertical transmission of the virus from parent to offspring.
-
Invertebrate iridescent viruses (IIV) infect inverte-brates, especially insect species that live in damp or aquatic habitats. Soil is believed to be an important environmental reservoir for IIVs. The effect of soil moisture and the presence of microorganisms on the persistence of CIV have been studied (57). The loss of activity of CIV in dry soil (6.4% moisture, −1000 kPa metric potential) was very rapid and was not studied beyond 24 h. However, soil moisture did not affect the rate of inactivation of virus in damp (17% moisture, −114 kPa metric potential) or wet soil (37% moisture, −9.0 kPa metric potential). In contrast, soil sterili-zation significantly improved the persistence of CIV activity, both in damp and wet soil. These figures represent half lives of 4.9 days for CIV in non-sterile soil, 6.3 days in sterilized soil, and 12.9 days for the control virus suspension. This study also concluded that extra-host persistence in soil habitats may be an important aspect of the ecology of IIVs.
Temperature affects the rate of deactivation as iridescent viruses are thermolabile and are inactivated within minutes at temperatures over 55℃ (19, 44). Aqueous suspensions of CIV showed a 10 fold reduction in titer after 50 days either at 4℃ or 25℃ (43).
Ultraviolet radiation has been used to deactivate CIV in laboratory studies (54), and exposure to solar UV light also resulted in a very rapid inactivation of CIV in water, with infectivity dropping by approximately nine logarithms after 24 hour exposure to sunlight (63). The rate of loss of activity of CIV was also confirmed by Hernandez et al (34). They tested the UV effect on CIV in aquatic habitats and found that direct sunlight causes rapid loss of activity (99.99% reduction).
The sensitivity of CIV to a selection of organic solvents, detergents, enzymes and heat treatment was assayed in Spodoptera frugiperda (Sf9) cells and by injection of inocula into larvae of Galleria mellonella. CIV was found to be sensitive to chloroform. Sen-sitivity (defined as a reduction of at least 1 log activity) was detected following treatment by 1.0 and 0.1% SDS, 1% Triton-X100, 70% ethanol, 70% methanol, 1% sodium deoxycholate, pH 11.1 and 3.0. No sensitivity was detected to 1% Tween 80, 1 M MgCl2, 100 mM EDTA, lipase, phospholipase A2, proteinase K, or trypsin at the concentrations tested. Viral activity was reduced by approximately 4 logs following heating to 70℃ for 60 min or 80℃ for 30 min (45).
Classification
Morphology and composition
Host range and pathology
Sensitivity
-
CIV virions contain a single AT rich linear ds DNA molecule that is circularly permuted and terminally redundant. Although the viral genome is a single linear double stranded DNA molecule the physical map is circular due to the circular permutation of the viral genome (21, 27). The dsDNA genome has been entirely sequenced using cycle sequencing by primer walking technology and found to comprise 212, 482 bp. The base composition of the viral genomic DNA sequence was found to be 71.37% A+T and 28.63% G+C. The CIV genome contains 468 open reading frames (ORFs) encoding for polypeptides ranging from 40 to 2432 amino acid residues; 50% (234 ORFs) of these ORFs are nonoverlapping (39). The linear double stranded DNA genome of CIV is not methy-lated in contrast to the majority of members from vertebrate hosts in the Iridoviridae family.
The genome of CIV contains extensive regions of short direct, inverted, and palindromic repetitive DNA sequences (26, 39). The coding function of these regions is unknown, although late transcripts were detected in the IIV-9 repeat region (15). ORFs 261R, 396L and 443R of CIV are found as large repetitive DNA elements in the CIV genome.
-
The codon usage of CIV was analyzed for the major capsid protein, (274L), the DNA dependent RNA polymerase (176R), the DNA polymerase (037L), and the thymidylate kinase (251L) genes. The most com-mon codons that are used for these four genes are generally the same (37). Phylogenetically related viruses normally show a similar pattern of codon usage, despite the fact that they have different hosts. Unfortunately none of the other IIVs have been analy-zed for codon usage for comparison with the results of CIV.
-
Promoter structures of a delayed early (DNApol) and a late (Major capsid protein, MCP) gene of CIV have been analyzed (49, 50). A small region (AAAAT TGATTATTTGTTTT), located between -19 and -2 relative to the mRNA start site, is responsible for promoter activity of the DNApol gene. Mutations in the AAAAT motif in this region have a major effect on promoter activity, showing that this motif is an essential part of the core promoter structure. Mutations at the downstream side have less effect (24th, 23th or 22nd nucleotides). The DNApol promoter as a whole does not show a common structure with other CIV genes. In a similar way, the critical promoter sequence of the ICR 169 (ATATCTCACAGGGGAATTGAA AC) one of two IE gene of FV3 is also not conserved in other FV3 genes (65). The critical AAAAT motif, on the other hand, was found in the 100 nt upstream of the putative translational start codons of several other putative CIV DE genes. MCP promoter activity is located between positions -53 and -29 relative to the transcription start site. To locate the MCP promoter sequence in a specific region will require further studies. In CIV transcripts the length of the 5' untrans-lated region is normally short. Transcription of DNApol initiated 35 nt upstream and that of MCP 14 nt upstream of the translational start site (50). CIV DNApol, helicase, and MCP transcripts most likely do not have polyA tails, because they could not be am-plified when an oligo dT primer was used in the RT step (50). This is also the case of FV3 transcripts.
-
The CIV genome contains 26 ORFs that are conserved across viruses in all the genera of the family (25). These genes should provide key factors for crucial elements for DNA replication, gene transcription, nucleocapsid assembly and virion architecture. These proteins may also be involved in essential interactions with the host, for instance to enter host cells, abrogate the host metabolism and establish infection. The genes that have been identified as essential players in viral transcription/DNA replication are 22L, 037L, 142R, 143R, 176R/343L, 184R, 282R, 349L, 355R, 369L, 376L, 428L, and 436L, and as protein processing and modifications are 98R, 179R/439L and 380R. The other core genes encode homologues of MCP, FV3 IE ICP46-like protein, ATPase, myristilated membrane proteins, hypothetical protein of Clostridium tetani, Erv1/Alr-like protein and two unknown proteins (274R, 393L, 075L, 118L/458R, 337L, 307L, 347L, 287R, 295L) and ORF 067R.
As the number of the sequenced iridovirus genomes increase, the lack of colinearity between many of these genomes may become apparent.
In addition to these conserved genes, other genes of interest in CIV include DNA ligase (205R), putative homologues of peptides with antibacterial (160L) and antifungal properties (ORF308L), putative bacterial mutator protein (414L), and DNA repair homolog proteins (369L, 100L) (39, 55, 59).
Genomic organization
Codon usage
Potential promoter sequences and transcription initiation sites
The conserved genes
-
The information that we know about the replication mechanism of iridoviruses comes mainly from studies of Frog virus 3 (FV3) and this has become the model for all iridoviruses. Although CIV is known as cyto-plasmic virus, it commences replication in the nucleus and completes it in the cytoplasm. The virus particle binds to a cellular receptor of the host cell. Following binding to host cell, enveloped virus is thought to enter via receptor mediated endocytosis, whereas naked virions uncoat by fusion at the plasma membrane. 096L of CIV is a good candidate for this binding protein since it has a fasciclin domain which may play a role in virion-cell adhesion. After penetration, the viral DNA is transported to the cell nucleus and DNA replication occurs here in units up to twice the genomic size. Early viral transcripts of two classes: immediate early (IE) and delayed early (DE) genes are synthesized here using input virion DNA as template (63). The DNA is then transported into the cytoplasm late in infection to form large concatemers up to ten-fold the genomic size (66). The concatemeric form of DNA is found within cytoplasmic viral assembly sites (AS). Late viral mRNA synthesis takes place either in the cytoplasm of infected cells or within AS in the cytoplasm. This can be catalyzed by a virus modified cellular polymerase or a novel RNA pol Ⅱ that is virally encoded. Concatamers are resolved into packaged lengths, possibly by a headful packaging approach. Virions exit the cell by budding or cell lysis.
The replication of CIV DNA in Cf124 cells has been studied (13, 22). Infections with 0.5 mg/mL active virus (V) resulted in initiation of CIV DNA replication by at least 6 h p.i. and accumulation of high levels of viral DNA by 24 h p.i. Low and declining levels of viral DNA were detected after infection with heat-inactivated virus (65℃, 30 min; VH). Infection in the presence of 5mg/mL aphidicolin (VA) showed levels of viral DNA only slightly above those observed with heat-inactivated virus. The declining levels of viral DNA in infections with heated virus (VH) suggested that viral DNA failed to replicate and was progressively degraded. Viral DNA levels in aphidicolin-treated infections (VA) were similar to parental DNA levels in infections with heated virus (VH), indicating that aphidicolin effectively inhibited viral DNA replication and possibly blocked degradation of viral DNA (22).
-
Previous studies on infected cell-specific poly-peptides and on CIV transcripts provided evidence for a temporal cascade subdividing the CIV mRNAs into three temporal classes: immediate-early (IE or α), delayed-early (DE, β), and late (L, γ) (3, 22). The temporal expression of three classes of CIV mRNA molecules during the course of infection suggests that both cis-acting DNA sequences and trans-acting regulatory factors interact at specific times post-infection to initiate transcription of the appropriate mRNAs. The immediate-early class contained 38 trans-cripts, appearing around 0.5 h.p.i., synthesized in the absence of de novo protein synthesis. It has previously been shown that purified CIV DNA is not able to start infection unless complemented with UV-irradiated virus particles (51), suggesting that for IE gene expres-sion in CIV, a virion-associated protein is required. The delayed-early class contained 34 transcripts, appeared around 3 h.p.i., are formed in the presence of DNA synthesis inhibitors, but not protein synthesis inhibitors, and require at least one earlier gene product for their expression. It is found that a protein of approximately 100 kDa, interacted with the active site of the DNApol promoter (delayed early gene) and may represent a transactivator protein (49). This protein is considered virus specific or virus-induced because in mock-infected control cells no binding was observed. Nuclear extracts prepared at different time points post-infection and in the presence or absence of protein synthesis inhibitors may help to determine the timing of expression of this 100 kDa protein in the expression process. The late class with 65 transcripts, appearing around 6 h.p.i. contained the full array of viral RNA formed only in the absence of inhibitors (22). The precise coding function of all these transcripts, however, was not clear from these studies.
Replication
Transcriptional regulation
-
CIV polypeptides have been studied by different groups until now (2, 20, 40). Barray and Devauchelle (2) reported that, after solubilization of CIV particles with SDS-β-mercaptoethanol, 16 polypeptides were resolved with molecular weights ranging from 7000 to 120.000, with a major polypeptide of 51.000, whereas after solubilization with SDS-urea 26 polypeptides were resolved with molecular weights ranging from 10.000 to 230.000. Cerutti and Devauchelle (12) detected structural proteins linked with disulfide bonds, since the importance of disulfide linked proteins for biological activities, structure and morphogenesis of viral particles is well known. However these studies have not revealed the total structural proteins of CIV. A recent LC-MS analysis of CIV revealed that there are a total of 55 polypeptides as structural proteins (Ince et al., manuscript in preparation).
-
Studies on interactions between CIV and cells have shown that inoculation of vertebrate or invertebrate cells with CIV rapidly leads to a massive formation of syncytia (9) and CIV rapidly inhibits cellular RNA, DNA and protein synthesis in permissive and non permissive vertebrate and invertebrate cell lines. The integrity of the viral genome is not required for inhibitory expression, since viral proteins solubilized from CIV by freezing and treatment with EDTA exhibit inhibitory properties similar to those of intact virions (10). This inhibition system is the same with the FV3, since it also depresses RNA, DNA and protein cellular synthesis under non-permissive condi-tions for virus replication (7, 31, 42).
In another study from Cerutti and Devauchelle, the CIV internal membrane was solubilized by octy-lglucoside treatment. This leads to the formation of vesicles. They showed that these reconstituted vesicles behave like the whole viral particle with respect the cell fusion and extensive inhibition of macromolecular syntheses in the host cells (11)
Recently it was shown that CIV virion protein extract (CVPE) induces mortality in neonate boll weevil larvae (6) and apoptosis in spruce budworm Choristoneura fumiferana and boll weevil cell cul-tures (56). It is also shown that apoptosis inhibition occurred under conditions permitting early viral expression (16). The candidate apoptosis genes are listed as ORF 157L, ORF 193R, ORF 332L, and ORF 284R. An interesting recent development comes from the work of Ince et. al. (36). CIV ORF 193R has been shown as a functional apoptosis inhibitor gene since it reduces significantly apoptosis in the presence of actinomycin-D. The 157L, 284R and 332L genes have not been tested, but it is anticipated that they are not functional as IAPs since they lack BIR domains. IAPs are characterized by the presence of one to three baculovirus inhibitor repeat (BIR) domains at the amino terminus and a C3HC4 RING finger domain at the carboxy terminus (reviewed by Clem, 17). All active iap genes determined until now, contain at least these two types of conserved domains, except the African swine fever virus IAP which contains a zinc instead of a RING finger (51).
-
High levels of homology have been detected bet-ween putative viral gene products of CIV and the corresponding viral proteins of lymphocystis disease virus of fish (LCDV). 183 ORFs of CIV genome posses homologs within the LCDV genome (39). However, no co-linearity was detected when the DNA nucleotide sequences and coding strategies of the CIV and LCDV genomes were compared to each other. Recently, Eaton et al. (25) performed a whole genome com-parative phylogenetic analysis of iridoviruses and found that the Iridovirus and Chloriridovirus genera are closely related to one another based on presence of orthologous genes. In contrast, the Megalocytivirus genus is divergent from the Iridovirus genus.
Cricket iridovirus (CrIV), isolated in 1996 from Gryllus campestris L. and Acheta domesticus L. (both Orthoptera, Gryllidae) was found to have between 95.98 and 95.18% average identity with selected seven genes to CIV at the nucleotide and amino acid level, respectively 38). The researchers concluded that CrIV must be considered to be a variant or a novel strain of CIV.














 DownLoad:
DownLoad: