-
The transactivator protein Tat of human immuno-deficiency virus-1(HIV-1) can strongly transactivate the HIV long terminal repeat (LTR) and is the most important regulator of viral gene expression and replication (7, 20). Hence, the Tat protein was considered an attractive target for antiviral therapy, and received much attention. Tat protein contains 86-104 amino acids depending on genotype, but three major functional domains are conserved: the N-terminal amphipathic α-helix domain, the cysteine-rich region which contains seven Cys residues and a third basic domain rich in Arg and Lys residues. This basic domain is responsible for interacting specifically with the transactivation response region (TAR) RNA (12, 21, 23). It has been proved that Tat is a multi-functional protein. One function is as an adaptor protein for cellular cofactors, many of which exhibit intrinsic enzymatic activities. Tat is also found to bind cyclin T1 through the highly conserved cysteine-rich region and recruit the cyclin-dependent kinase 9 (CDK9) to elongate HIV transcripts (1). Tat and cyclinT1 bind TAR cooperatively, and then induce phosphorylation of the C-terminal domain of RNA polymerase Ⅱ by CDK9. Among the Tat-binding proteins, there are a number of transcriptional coactivators with intrinsic histone acetyltransferase activity, e.g p300/CBP, p300/CBP-associated factor (PCAF), TAFⅡ250 and Tat-interacting protein 60 (Tip60) (1). Since Tat also induces remodeling of a single nucleosome (nuc-1) positioned at the HIV promoter, it was proposed that Tat stimulates transcriptional elongation of HIV both by increasing the intrinsic activity of the RNA polymerase Ⅱ complex to elongate more efficiently, and by recruiting histone-modifying enzymes to remodel the elongation block caused by nuc-1.
Despite its nuclear localization and function and the lack of any secretory signal sequence, Tat was found to be secreted by HIV-infected cells and to be taken up by affected neighbouring cells. Therefore, Tat is regarded as an important contributor to the pathogenesis of AIDS. The influence of this exogenous "bystander" effect on both infected and uninfected cells was thought to be mediated by its protein transduction domain (PTD). In fact, Tat can also fulfill the function as an exogenous factor, through interacting with a number of cellular membrane receptors including fibronectin, adhesion proteins and other cellular receptors (5, 10, 18, 24, 26) and then being internalized by means of endocytic activity (12). However, the exogenous activity and function of Tat is poorly studied due to difficulties with heterogeneously expressing this protein in E.coli bacteria. The preparation of Tat protein was assumed to be hampered by its high cysteine content and its susceptibility to oxidation leading to the formation of stable multimers (11). The successful cases of production of recombinant Tat are mainly restricted to the first exon of the tat gene (13, 19) or synthetic peptides (2). It was suggested that Tat protein exists in inclusion bodies and monomer forms as expression of Tat in E.coli (16) and the yield of purified product was relatively low (9). In this article, we cloned a full-length tat gene and compared the codon usage bias between eukaryotic Homo sapiens and prokaryotic E.coli. Results indicate that there are at least 14% rare codons in the Tat protein for E.coli. Therefore, the high ratio of rare codons might be the major reason for the poor expression level of Tat protein in E.coli. Just by changing the host strain from BL21(DE3) to Rosetta, which is supplemented with six tRNAs for Arg, Leu, Ile and Pro, the expression level of Tat increased dramatically. The achievement of effective expression and purification of the full-length, soluble dimer of Tat from a native gene, instead of a synthetic one, has paved the way to implement a method to fully investigate the exogenous function of HIV-1 Tat.
HTML
-
Primers were synthesized by Sunbiotech Company, Beijing, China. Restriction endonuclease and T4 DNA ligase were purchased from New England Biolabs. High fidelity and hot start polymerase KOD was purchased from TOYOBO, Japan. GSTrap FF column was from Amersham Biosciences and Ni-NTA His-Bind Resin was from Qiagen. Mouse monoclonal anti-His6 tag antibody was from Novagen.
-
To clone the full length tat gene, total RNA was extracted from the leucopheresis peripheral blood cells of an AIDS patient using Trizol reagent (Invitrogen). RT-PCR was performed to get the total cDNA. The two exons of the tat gene were separately amplified by PCR. Briefly, primer1 & primer2 were used to amplify the first exon and primer3 & primer4 were used to amplify the second exon. Primer sequences are as follows, Primer 1: 5'-gtgaattctcgacagaggagagcaagaa-3' (the EcoR Ⅰ restriction site is underlined); Primer 2: 5'-ctctgggctgggaagcgggttgctttgatagagacgctt g-3'; Primer 3: 5'-caagcgtctctatcaaagcaacccgcttccca gcccaga-3'; Primer 4: 5'-gtgtcgaccgtcccagaagttccacaat-3' (the Sal Ⅰ restriction site is underlined). The down-stream primer of exon 1 (Primer 2) and the up-stream primer of exon 2 (Primer 3) are overlapped, the PCR fragments of exon 1 and exon 2 were mixed and annealed, and amplified for 10 cycles without primers, then the full length ORF of the tat gene was amplified by PCR using primer1 and primer4.
-
For construction of the tat expression vector with a GST tag at its N-terminus, tat cDNA was amplified by PCR using the full length cDNA of tat mentioned above as template. PCR was performed by using highly fidelity hot start polymerase KOD. PCR primers were as follows: primer 5, 5' gcggatccatggagccagtagatcctag 3'; and primer 6, 5' gcctcgagttaatctctc ggatctgtctttg 3' (BamH Ⅰ and Xho Ⅰ restriction sites are underlined). PCR conditions were as follows: denaturation at 94℃ for 30 sec, annealing at 60℃ for 30 sec and extension at 72℃ for 30sec for 32 cycles. PCR product and expression vector pGEX5T-1 were digested with BamH Ⅰ and Xho Ⅰ respectively, and the tat gene was inserted into pGEX5T-1 vector. For the construction of pET22b-tat which fused with a His(6)tag at its C-terminus, the tat gene was amplified using the following primers: primer7, 5'gcgaattcatggagccagtagatcctag3'; and primer 8, 5' gcctcgagatctctcggatctgtctttg3' (EcoR Ⅰ and Xho Ⅰ restriction sites are underlined). The PCR product was inserted into pET22b expression vector between the EcoR Ⅰ and Xho Ⅰ restriction sites. The recombinant expression vectors pET22b-tat and pGEX5T-tat were confirmed by both endonuclease digestion and DNA sequencing.
For construction of the tat expression vector with a GST tag at its N-terminus, tat cDNA was amplified by PCR using the full length cDNA of tat mentioned above as template. PCR was performed by using highly fidelity hot start polymerase KOD. PCR primers were as follows: primer 5, 5' gcggatccatggagccagtagatcctag 3'; and primer 6, 5' gcctcgagttaatctctcggatctgtctttg 3' (BamH Ⅰ and Xho Ⅰ restriction sites are underlined). PCR conditions were as follows: denaturation at 94℃ for 30 sec, annealing at 60℃ for 30 sec and extension at 72℃ for 30sec for 32 cycles. PCR product and expression vector pGEX5T-1 were digested with BamH Ⅰ and Xho Ⅰ respectively, and the tat gene was inserted into pGEX5T-1 vector. For the construction of pET22b-tat which fused with a His(6)tag at its C-terminus, the tat gene was amplified using the following primers: primer7, 5'gcgaattcatggagccagtagatcctag3'; and primer 8, 5' gcctcgagatctctcggatctgtctttg3' (EcoR Ⅰ and Xho Ⅰ restriction sites are underlined). The PCR product was inserted into pET22b expression vector between the EcoR Ⅰ and Xho Ⅰ restriction sites. The recombinant expression vectors pET22b-tat and pGEX5T-tat were confirmed by both endonuclease digestion and DNA sequencing.
-
The expression plasmids pET22b-tat, pGEX5T-tat and control plasmid pGEX5T-1 were transformed into E.coli BL21(DE3) and Rosetta-gami B(DE3). For the BL21(DE3), a single colony of transformants was inoculated in fresh liquid LB medium containing 100mg/L ampicillin (for BL21 transformant), or containing 34mg/L chloramphenicol, 16mg/L kanamycin, 12.5mg/L tetracycline and 100mg/L ampicillin (for the Rosetta transformant). When the culture reached the mid-log phase (OD600≈0.6), IPTG was added to induce the expression of target protein at a final concentration of 0.5mmol/L. Three hours later, cultures were collected and resolved in 15% SDS-PAGE. The SDS-PAGE gel was scanned after Coomassie blue staining.
-
The purification of Tat-his was performed according to the manufacturer's instruction for Ni-NTA His-Bind Resin. Cells were harvested by centrifugation at 6000r/min for 10min. 10 grams of cell pellets from 1 L culture were re-suspended in 50mL 50mmol/L sodium phosphate, 0.3 mol/L NaCl, pH8.0 and lysed by ultrasonic liquid processor (Sonxi, China) for 30min at 4℃. The supernatant was collected by centrifugation at 12 000 ×g for 20min.The resulting supernatant was filtered with 0.45 μm membrane and loaded onto Ni2+ affinity column pre-equilibrated with buffer (50mmol/L sodium phosphate, 0.3 mol/L NaCl, 10mmol/L imidazole, pH8.0). After washing with buffer (50mmol/L sodium phosphate, 0.3 mol/L NaCl, pH 8.0, 10mmol/L imidazole), the Tat-hiswas eluted with the buffer (50mmol/L sodium phosphate, 0.3 mol/L NaCl, 250mmol/L imidazole, pH 8.0, ). Eluted Tat-hisprotein was analyzed with SDS-PAGE, and the purity and concentration was determined with the Quantity One software package (Bio-Rad) and the Bradford method respectively. The final purity of Tat-hiswas over 85% and concentration was 0.5μg/mL.
The purification of GST and GST-tat was performed according to the manufacturer's instruction for the GSTrap FF column purchased from Amersham Biosciences.
-
Protein samples were run in 15% SDS-PAGE, and transferred to nitrocellulose membrane. The membrane was blocked with 5% non-fat milk in TBS-T (20mmol/L Tris-HCl, 137mmol/L NaCl, 0.1% Tween-20) and detected with anti-His antibody (Novagen) or anti-GST antibody (Sigma) and then peroxidase-conjugated secondary antibody. The strip was processed with an enhanced ECL Western blotting kit (Santa Cruz). Control experiments were carried out with empty vector or with normal mouse Ig G antibody.
-
Human kaposi's sarcoma cell line TE671 was cultured with Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. The activity of transactivation of the recombinant Tat protein was assayed by using Tat transcription activating reporter vector KB/SP1LTR-nlacZ, kindly provided by Dr. L. J. Cheng (Department of Molecular Genetics and Microbiology, University of Florida), in which the reporter lacZ gene expression is driven by the LTR element activated by the tat protein. TE671 cells were transfected with plasmid KB/ SP1LTR-nlacZ using Lipofectamine 2000 (Invitrogen). At 48 h after transfection, control and transfected cells cultured on glass coverslips were rinsed with PBS, subjected to fixation with 1%(v/v) paraformaldehyde and 0.2%(v/v) glutaraldehyde for 30min. After washing two times with PBS, the glass coverslips were stained with 5-bromo-4-chloro-3-indolyl β-D-galactoside (X-Gal) staining solution (PBS containing 4mmol/L K3Fe (CN)6, 4mmol/L K4Fe(CN)6·3H2O, 2mmol/L MgCl2, and 0.4mg/mL X-Gal) for 3 h.
It has been reported that the basic domain of the Tat protein possesses the ability to traverse biological membranes efficiently in a process termed protein transduction. Although the mechanism is unknown, transduction occurs in a receptor-and transporter-independent fashion that appears to target the lipid bilayer directly. Furthermore, HIV-1 Tat proteins have recently been shown to serve as carriers to direct uptake of heterologous proteins into the cells in vitro and in vivo.
-
Protein spots on the electrophoresis gel were excised and transferred into 1.5 mL Eppendorf tubes. Gel chips were destained with solution containing 50mmol/L ammonium bicarbonate, 50% acetonitrile at pH8.0. After hydrating with acetonitrile and drying, the gel chips were rehydrated in a minimal volume of trypsin solution (10μg/mL in 25mmol/L NH4HCO3) and incubated at 37℃ overnight. Recovered peptides were prepared for MALDI-TOF-MS by mixing 0.5μL of the peptides mixture with 0.5μL of 5 mg/mL α-Cyano-4-hydroxycinnamic acid (CHCA) and allowing the droplet to dry on the MALDI sample plate.
The mass spectra were recorded by using a time-of-flight delayed extraction MALDI mass spectrometer (Bruker Autoflex). Samples were mixed in an Eppendorf tube with equal volume of matrix solution. A solution of α-Cyano-4-hydroxycinnamic acid (CHCA) was prepared at a concentration of 15mg/mL in 2:1 (v/v) acetonitrile/0.1%TFA. 2 μL of the mixture was applied to a gold-plated sample holder and introduced into the mass spectrometer after drying. The spectra were obtained in the reflectron mode by summing 200 laser shots with an ion source voltage 1 of 19 kV, ion source 2 of 16.27kV, 100 ns delay and the low mass gate at m/z=600. Monoisotopic peptide masses obtained from MALDI-TOF were queried against entries for protein databases in NCBI using the protein search program, Mascot (Matrix Science Ltd, London). Molecular weight of Tat was determined by MALDI-TOF using the same method described above except it was not digested.
Materials
Cloning the full length tat gene
Construction of tat expression vectors
Expression of Tat protein
Purification of Tat protein
Western blotting analysis
Cell culture and reporter gene assay
The protein identification by MALDI-TOF MS
-
The full length coding region of the tat gene was cloned as described in the methods (GenBank accession number: EU001659), and sequence analysis revealed that there exists a greater diversity in the sequence as compared to the Tat proteins from other HIV virus strains (Fig. 1). However, its cysteine-rich domain comprising seven cysteines is conserved.

Figure 1. Comparison of Tat protein sequence we cloned with other different virus strains. The GenBank accession numbers are as follows: BH10 (M15654, K02008, K02009, K02010), CNHN24 (AY860947), 97CNGX (AAG38928) and the sequence we cloned 03CHHN (EU001659).
In the beginning, we tried to express this Tat protein from the E.coli BL21 (DE3) strain, but found that the this heterogeneous tat gene hardly expressed in E.coli BL21 (DE3). It has been reported that some eukaryotic genes are hard to express during heterologous gene expression, and the high cysteine content was assumed to be a key reason for this inhibition (11). Therefore, we compared the codon usage bias difference between Homo sapiens and E.coli using the Graphical codon usage analyzer (http://gcua.schoedl.de/seqoverall_v2.html). Comparison results indicated that there is a 40.5% mean difference (Fig. 2) mainly attributed to codons for Arg, Leu, Ile and Pro (Table 1). Among the E.coli rare codons encoded by tat, there are 9 of 9 for Arg, 1of 4 for Leu, 1 of 2 for Ile and 3 of 11 for Pro (Table 1).
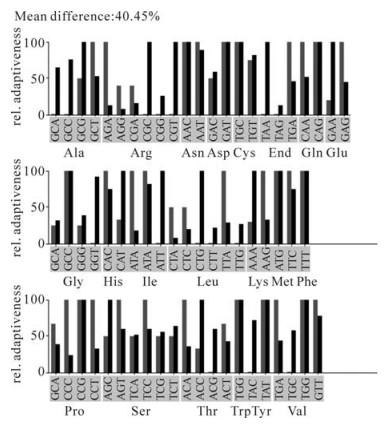
Figure 2. Codon usage of HIV-1 tat gene between Homo sapiens and E.coli. codon usage analysis was performed in the Graphical codon usage analyzer website: http://gcua.schoedl.de/seqoverall_v2.html. Red bars means codon relative adaptiveness in Homo sapiens and black bars means codon relative adaptiveness in E.coli. Relative adaptiveness was derived from the codon usage frequency values and in positive correlation with it.

Table 1. The rare codons of tat gene for E.coli
-
To express Tat-his6 and GST-tat protein, pET22b-tat-his6 and pGEX5T-tat constructs were transformed into BL21 (DE3). After induction with IPTG, the lysate was subjected to SDS-PAGE and western blot analysis. As Fig. 3 shows, neither Tat-his6 nor GST-tat can be expressed in BL21 (DE3), no signal of Tat protein can even be detected by the western blot. As mentioned above, the rare codons could be key contributors to the inhibition of tat expression. Indeed, at least 14% (14 of 101) of the codons were rarely used in E.coli. Notably, all of the codons for Arg were rarely used in E.coli (Table 1). Accordingly, we then used the Rosetta strain, which was supplemented with six tRNAs for Arg, Leu, Ile and Pro compared to the BL21 (DE3). After induction with IPTG, the lysate was analyzed with SDS-PAGE and Western blot. As shown in Fig. 3, both Tat-his6 and GST-tat were expressed at high levels in the Rosetta strain, subsequent Western blot analysis also confirmed this expression. Further analysis revealed that all the expressed Tat-his6 and GST-tat fusion protein were soluble despite the fact that the induction was performed at 37℃.
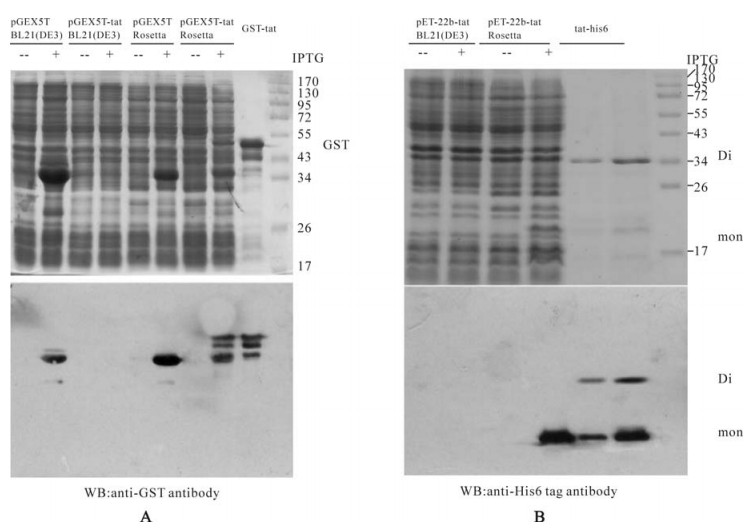
Figure 3. A: Expression of GST (pGEX5T), GST-tat (pGEX5T-tat) fusion protein in E.coli BL21 (DE3) and Rosetta strains and Western blot diction with anti-GST antibody. B: Expression of Tat-his6 fusion protein (pET22b-tat) in E.coli BL21 (DE3) and Rosetta strains and Western blot with anti-His6 antibody.
-
To further characterize the recombinant Tat, trypsinized or non-trypsinized proteins were subjected to MALDI-TOF-MS analysis. Firstly, the molecular weight of purified Tat-his6 was calculated by MALDI-TOF-MS. As shown in Fig. 4, the molecular weight of Tat (12548.843, Fig. 4 A) is closer to the theoretical value (12599.30) when protease inhibitor cocktail (Roche) was added to the lysate during purification and the mass was measured immediately afterwards. On the contrary, the molecular weight value (11039.256, Fig. 4 B) is much smaller than the theoretical one when protease inhibitor was not added to the lyaste during purification and measured 48 h after purification at 4℃, which means the protein lost more than ten amino acid residuals. Secondly, we measured the peptide mass fingerprint (PMF) of recombinant Tat by MALDI-TOF-MS and predicted the PMF of Tat using the peptide-mass tool (http://expasy.org/tools/peptide-mass.html). There was some difference between measured and predicted PMF (Table 2, Fig. 4C). We believe that this difference can be attributed to the diversity between different strains. In fact, the database search (www. matrixsciences.com) revealed that measured PMF can be matched with other mutants of Tat (data not shown).
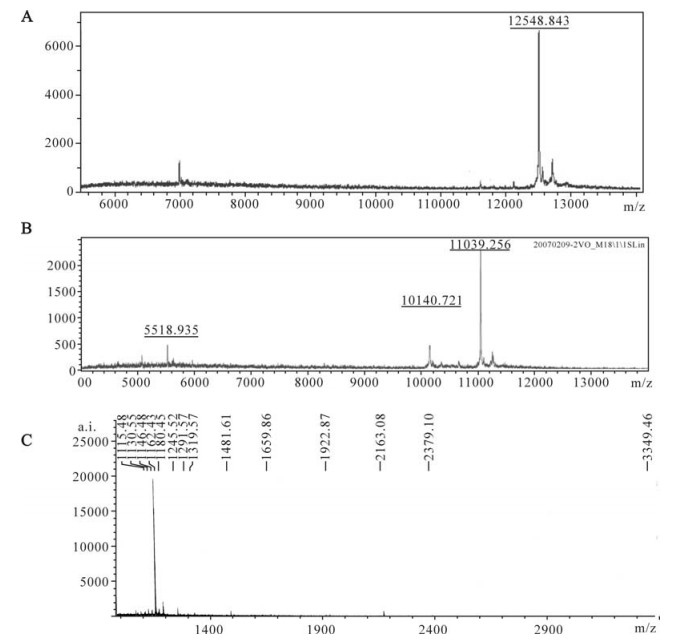
Figure 4. MALDI-TOF-MS analysis of recombinant Tat protein (molecular determination). A: The molecular weight of recombinant Tat protein determined by MALDI-TOF-MS immediately after purification. B: The molecular weight of recombinant Tat protein determined by MALDI-TOF-MS after 48h storage at 4℃ post-purification. C: The peptide mass fingerprint (PMF) of recombinant Tat protein as analyzed by MALDI-TOF-MS.

Table 2. The predicted and measured peptide mass fingerprint
-
As shown in Fig. 3B, two bands (MW 14kDa and 25kDa) of the purified Tat protein appear in SDS-PAGE gel. Western blot proves that both bands correspond to Tat-his6 proteins, which means that the larger one is the Tat dimer and the smaller is the monomer. The β-mercaptoethanol is commonly added to the loading buffer and functions as a reductant and breaks the disulfide bond between the two molecules. The results above indicate that Tat forms a dimer not by a cysteine disulfide bond but by another mechanism.
-
To further analyze the biological activity of this recombinant Tat, reporter plasmid KB/SP1LTR-nlacZ was transfected into the TE671 cell. The purified Tat protein was added immediately or after 48h of storage at 4 ℃into the cell culture to analyze the activity. In this assay, the expression of lacZ gene is driven by the Tat-activated HIV-1 LTR element, and it can be detected by means of X-gal staining. Staining results indicated that the recombinant Tat protein can efficiently activate the transcription of the target reporter gene, even after 48h storage at 4℃ post-purification (Fig. 5B).
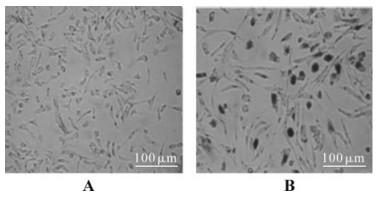
Figure 5. Detection of the transactivation activity of the recombinant Tat protein. A: TE671 cells were transfected with reporter plasmid KB/SP1LTR-nlacZ, but without the treatment of recombinant Tat protein. B: TE671 cells were transfected with reporter plasmid KB/SP1LTR-nlacZ, and treated with the recombinant Tat protein at final concentration of 100 ng/mL.
Sequence and codon analyses of newly cloned tat gene
Tat fusion protein expressed at high level in Rosetta but not in BL21 (DE3) E.coli
Characterization of recombinant Tat
Recombinant tat does not form dimers by cysteine disulfide bonds
Confirmation of transactivation activity of the recombinant Tat protein
-
Obtaining an intact Tat protein which exhibits natural properties is an important achievement for the functional study of the HIV Tat protein and the mechanism elucidation of AIDS pathogenesis. There are some reports regarding the expression of tat in E.coli, but these successful cases were mainly restricted to the first Tat exon (13, 19) or to synthetic peptides (2). There are several obstacles impeding the effective expression of the HIV tat gene in E. coli, e.g. the yield of purification is relatively low (9) or the expressed tat only appears in the form of an inclusion body and monomer tat (16). The difficulty of achieving heterologous expression of HIV-1 tat in E.coli hampered the complete study of its exogenous activity in cells. In this report, we cloned a full-length tat gene from an HIV-1 infected patient -the sequence has been submitted to GenBank (accession number: 925024). Here we demonstrated that the HIV-1 tat gene cannot be induced to express in E.coli strain BL21 (DE3). Therefore, we performed codon usage bias analysis and the results demonstrated that at least 14% of codons of HIV-1 tat are rarely used in E.coli, including codons for Arg, Leu, Ile and Pro. It has been reported that the protein expression of those genes containing rare codons can be dramatically improved when the cognate tRNA is increased within the host (4, 22, 25). Most of the reports focused on the rare codons AGG (22), AGA (6, 22) for Arg, ATA (8, 27) for Ile, CTA (15, 17) for Leu, and CCC for Pro. Based on this information, we presumed that the major cause inhibiting the expression of HIV-1 tat gene in E.cloi was the elevated presence of these rare codons, especially the consecutive Arg rare codons (at positions: 49th, 52th, 53th, 55th, 56th and 57th) which cause translational stalling and termination during protein biosynthesis. Hence, by using modified BL21 (DE3) E.coli, named Rosetta-gami B (DE3) (Novagen), which supplied tRNAs for the codons AUA, AGG, AGA, CUA, CCC, and GGA (14), this HIV-1 tat gene was dramatically induced into gene expression.
One laboratory reported that Tat can only be expressed as inclusion body and monomer in E.coli (16) and another report showed that Tat can form metal-linked dimers (11), perhaps the metal ion Zn2+ or Cd2+. Our results demonstrated that recombinant Tat was soluble and formed dimers in a way which, similar to its native state, does not involve the formation of cystine disulfide bonds. Measurement of molecular weight of recombinant Tat at different time points revealed that recombinant Tat is unstable due to its vulnerability to protease or its high basic amino acid residual content. The underlying mechanism should be investigated later. Reporter gene activation confirmed biological activity of Tat, even in the truncated form, is caused by partial degradation. In conclusion, our improved strategy for efficient expression and purification of soluble and dimer Tat has paved the way for in depth study of the exogenous biological activity of the HIV-1 Tat protein. These findings will be very helpful seeking novel methods for AIDS treatment via targeting the Tat pathway in the near future.














 DownLoad:
DownLoad: