-
In eukaryotic cells, the translational machinery and metabolic systems of the cytoplasm are separated from the genetic material and transcriptional machinery of the nucleus by the nuclear envelope. Transport of macromolecules larger than 60 kDa between the nucleus and the cytoplasm is an active, energy-dependent and essential cellular process that is mediated by sequence-specific motifs, nuclear localization signals (NLS), and nuclear export signals (NES) [3]. This review summarizes the mechanisms having been identified so far for the nucleocytoplasmic transport of macromolecules and viral proteins.
HTML
-
The bidirectional transport of macromolecules between the nucleus and cytoplasm is a selective process that occurs exclusively through nuclear pore complexes (NPC) [1, 10]. The NPC, is a 125-MDa macromolecular assembly of 50 to 100 polypeptides [4], consists of a cylindrical structure with an 8-fold symmetry and a central diameter of about 10 nm [3]. The NPC allows the passive diffusion of small molecules, including ions, metabolites, and globular proteins of up to ca. 60 kDa [9], without energy consumption. However, transport of larger proteins between the cytoplasmic and nuclear compartments is an active process and facilitated by specific soluble carrier proteins that are collectively referred to as "karyopherins", with "importins" and "exportins" [20]. The karyopherin family comprises about 14 known members in yeast and 19 in humans [35]. Many transport receptors are collectively called "β-karyopherins" because the majority of them are members of the importin β superfamily [20]. Although cargo proteins can bind directly to β-karyopherins, the interaction between the β-karyopherin and the cargo is mediated by the importin α during classical nuclear import [20].
The energy for nuclear transport is provided by Ran (a small Ras family of GTPases) [30], which cycles between a GTP-and a GDP-bound state. An asymmetric distribution of Ran-GTP and Ran-GDP between the nucleus and the cytoplasm controls the cargo import and export, and this gradient is maintained by various Ran associated regulatory factors [15]. Ran-GTP is concentrated in the nucleus, while Ran-GDP is concentrated in the cytoplasm, and Ran can impart directionality to nuclear transport by acting as a molecular switch that controls the binding and release of cargo for the compartmentalization [15, 34].
In import pathways, Ran-GTP and substrates bind Kap s competitively, allowing substrate binding in the cytoplasm and Ran-GTP-mediated release in the nucleus [21]. In the cytoplasm, a cargo molecule containing an NLS binds to an importing karyopherin (importin) in the absence of Ran-GTP. The import complex docks at the cytoplasmic side of an NPC and is then translocated into the nucleus [29]. Once inside the nucleus, imported Kapβs bind the small GTPase Ran-GTP and release their substrates [35]. By interaction with the active, guanosine 5′-triphosphate (GTP)-loaded form of the small guanosine nucleotide-binding protein Ran, i.e. with Ran-GTP, the import complex is dissociated, and the cargo is set free to search the nuclear contents for a specific binding partner [29]. In contrast, in export pathways, Ran-GTP, substrates, and Kapbs bind cooperatively, resulting in substrate binding in the nucleus and release in the cytoplasm as the Ran bound nucleotide is hydrolyzed [21]. Whereas a cargo containing an NES forms a trimeric export complex with exporting karyopherin (exportin) and Ran-GTP in the nucleus. The export complex docks at the nuclear side of an NPC and is translocated into the cytoplasm [29]. In the cytoplasm, Ran-bound GTP is hydrolyzed, inducing the dissociation of the complex. Importing and exporting karyopherins, although quite large (~100 kDa), can pass through the NPC freely [29]. The diameter of the NPC can increase to some 30 nm in order to allow larger complexes come through. Once transport has been completed, the cargo is released and the transporters are recycled [3], while the import of Ran into the nucleus is facilitated by NTF2, a nuclear transport receptor belonging to the nuclear export factor (NXF) family of nuclear transport receptors [3, 29].
Full understanding of nucleocytoplasmic transport is important for both medical science and biotechnology. The role of nucleocytoplasmic transport plays in the biogenesis of pathological conditions and the possibilities for new therapeutic strategies have been intensively studied. In addition, the NPC provides an intriguing paradigm of a molecular machine to screen native protein molecules with high efficiency [29].
-
Many proteins must enter the nucleus to fulfill their physiological roles [4].The best understood system for the transport of macromolecules is the classical nuclear import pathway, which was first proposed when a nuclear targeting signal in the large T antigen of the simian virus 40 (SV40) was characterized [16]. Since then, several pathways for nucleocytoplasmic transport have been described [20].
NLS motifs consist of grouped positively charged amino acids and can be mono-or bipartite. The 'pat4' NLS consists of a continuous stretch of four basic amino acids (lysine or arginine) or three basic amino acids associated with histidine or proline. The 'pat7' NLS starts with a proline and is followed within three residues by an amino acid sequence containing three basic residues out of four, such as PKKKRKV [32]. The 'pat4' NLS and the 'pat7' NLS belongs to monopartite signals. The third type of NLS, known as a ''bipartite'' motif, consists of two groups of 2-3 basic amino acids separated by about 10 residues [14], such as KRPAATKKAGQAKKKKLD [7]. This information has been incorporated into computer programs to identify NLS motifs, but cannot always successfully predict the localization of a protein to the nucleus [32]; proteins without an NLS can be co-transported with other nuclear proteins and proteins that contain NLS sequences may remain cytoplasmic, especially if the NLS sequence is blocked or buried within the protein [31]. It is also possible that the sequence is not linear, in which case the basic residues may form a conformational epitope on the surface of the protein [3]. Furthermore, amino acid replacement analyses revealed that the consensus basic patterns of the classical NLS are not essential for activity, thereby generating more unconventional patterns, including redox-sensitive NLS. These results explain the causes of the NLS diversity [17]. Although the consensus sequences of the classical NLS have been defined, there are still many NLS that do not match the consensus rule and many nonfunctional sequences that match the consensus. It is reported that M9 is a novel type of nuclear localization domain as it does not contain sequences similar to the classical basic-type NLS, which is necessary and sufficient for nuclear localization [33]. Rather, it is a new NLS, PY-NLS, a C-terminal consensus sequence R/K/H-X(2-5)-P-Y (where X is any residue), that recognized by Kapβ2 [21]. Many studies have shown that the M9-NLS peptide is both necessary and sufficient for Kapβ2 mediated nuclear import [33].
In our previous studies, the subcellular localization of bovine herpesvirus type 1 (BHV-1) VP8 and VP22 have been characterized and their nuclear localization signals had also been identified. The results indicated that VP8 contained a pat4 NLS and VP22 contained a functional nonclassical NLS [36, 37]. Recently, we have identified and confirmed a functional NLS and nucleolar localization signal (NoLS) of BHV-1 infected cell protein 27 (BICP27) [13].
NLS confers interaction with members of the importins. There are two types of importins, α and β, both of which are importin β super-family numbers. Importin α serves as an adaptor that links cargos and importin β1 and recognizes NLS within the cargos [3]. Importin α recognizes two classes of classical NLS: monopartite NLS and bipartite NLS [20]. Once the cargo is bound by importin α, the complex can be recognized and bound by importin β that subsequently binds to the fibrils of the NPC and is responsible for the actual translocation [2]. Nuclear import of proteins is generally initiated by the formation of a ternary complex with importin α, importin β1, and a cargo, where importin β1 docks the complex to the NPC to release the cargo into the nucleus through the binding of Ran-GTP to importin β1 [17]. This is not the only way molecules can be transported into the nucleus [25] but it is the most common way. Importin α has several isoforms that recognize different variants of NLS but they are all recognised by a single importin β1. Therefore, these isoforms could be considered as adaptors for different signal transmission to the importin β1 molecule [6]. In the "classical" or "canonical" import pathway, an NLS-containing cargo is bound first by importin α. In the second step, the cargo-importin-α complex is bound by importin β1 and only the trimeric complex is translocated into the nucleus finally (Fig. 1) [29].
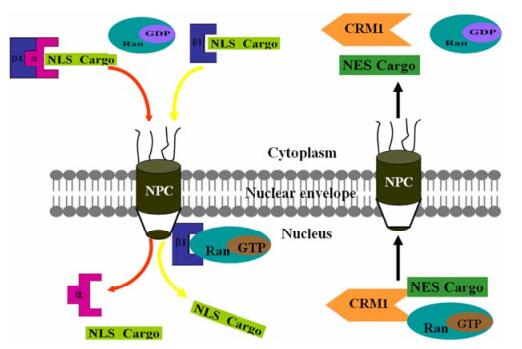
Figure 1. Simplified model of importin/exportin-mediated nucleocytoplasmic transport of proteins. In the cytoplasm, importin-α (α) binds to the NLS-containing cargo protein and forms a ternary complex with importin-β1 (β1) to enter into the nucleus through the NPC. Some NLS cargo can directly bind to importin-β1. Once within the nucleus, binding of Ran-GTP to importin-β1 elicits a conformational change resulting in cargo release into the nucleus. For nuclear export, Ran-GTP stimulates binding of exportin CRM1 to the NES cargo in the nucleus and the complex is exported to the cytoplasm, where hydrolysis of Ran-GTP to Ran-GDP.
Each import Kapβ appears to bind distinct sets of substrates, suggesting that each Kapβ recognizes different NLS. However, large sequence diversity among various substrates has hindered identification of NLS for most Kapβs, and it remains extremely difficult to predict NLS in candidate import substrates [21]. It is reported that six different NLS classes specifically bind to distinct binding pockets of importin α [17]. The well-defined consensus patterns and properties of importin α-dependent NLS will provide useful information for NLS identification.
The nuclear import pathways benefit viral pathogens to achieve efficient nuclear import of both viral particles during the initial stages of infection and individual viral proteins following their synthesis in the cytoplasm [4].
-
NES have an important regulatory function in the subcellular localization of viral proteins, and also have an impact on transcription and other nuclear processes [19]. Although several classes of NLS have been characterized, only one class of NES, the leucine-rich NES, is known at present [8]. The 10-15-residue leucine-rich NES were first characterized in the human immunodeficiency virus type 1 (HIV-1) Rev protein and cyclic-AMP-dependent protein kinase inhibitor [8]. Several mutagenesis and computational studies have identified a consensus sequence ϕ-X(2-3)-ϕ-X(2-3)-ϕ-X-ϕ, in which ϕ refers to Lys, Val, Ile, Phe or Met, X represents any amino acid, and the numbers in parentheses denote the number of repeats. Although this sequence matches most known leucine-rich NES, it is so broadly distributed that it also exists in most helix-containing proteins [8]. Prolines situated between the hydrophobic residues can disrupt the function of nuclear export [8]. Moreover, NES that do not belong to the family of leucine-rich NES have also been described for various proteins, as demonstrated by the atypical NES in the hepatitis D antigen HDAg-L [22].
Direct interaction of proteins bearing leucine-rich NES with the nuclear export factor chromosome region maintenance 1 (CRM1), also known as exportin-1, is very well documented and essential for the active nuclear export of these proteins [11, 12, 27]. CRM1 mediates nuclear export of hundreds of proteins through the recognition of the leucine-rich NES. The NES recognized by CRM1 is the consensus sequence ϕ-X(2-3)-ϕ-X(2-3)-ϕ-X-ϕ [3]. The large hydrophobic residues are the most conserved in NES which suggests that the interaction with CRM1 is mediated through these hydrophobic residues [19]. CRM1, contains armadillo repeats, belongs to the importin β super family, and is the regular exportin for RNAs, proteins and ribonucleoproteins complexes. Like importins, there exist exportins that directly recognize various NES and then bind to the second exportin molecule that interacts with the nuclear pore [3].
CRM1 cannot leave the nucleus with its cargo without the assistance of Ran [3]. It's believed that Ran-GTP and leucine-rich NES bind cooperatively to CRM1 upon formation of a ternary CRM1-Ran-GTP-NES complex (Fig. 1) [11, 28]. Moreover, the NES substrate RanBP3/Yrb2p functioning as an essential export cofactor, enhances the affinity of CRM1 for Ran-GTP [19].
Recently, a study showed that CRM1 binds an export substrate snurportin 1 (SNUPN) and Ran-GTP cooperatively in the nucleus [8]. Multipartite recognition in SNUPN may be used more generally to increase CRM1 binding beyond the usually low affinity leucine-rich NES [8]. The crystal structure of the CRM1-SNUPN complex has been presented [8]. This trimeric complex is translocated from the nucleus to the cytoplasm through the NPC [9]. Soon after the complex has left the nucleus, GTP is hydrolysed by Ran through interaction with Ran-GAP (Ran GTPase Activating Protein) which is associated with cofactors RanBP1 and RanBP2 (Ran Binding Proteins) that are localized on the fibrils on the cytoplasmic side of the NPC. After hydrolysis of GTP, the conformation of Ran changes and the complex falls apart and the exportins and Ran-GDP go back to the nucleus leaving the cargo remain in the cytoplasm [3].
Several ways of regulating NES-dependent export have been reported, such as masking/unmasking of NES phosphorylation and even disulfide bond formation resulted from oxidation [19].
Leptomycin B (LMB), a metabolite that disrupts the interaction of NE with the CRM1 receptor by binding to a cysteine residue, is localized in the central domain of the receptor [18]. Antifungal antibiotic LMB inhibits nuclear export by alkylating Cys 528 of human CRM1 [18]. It is unclear whether LMB blocks substrate binding sterically or by a conformational change [8]. Besides, LMB is used to identify CRM1 involved nuclear export pathways. Although the CRM1-dependent transport is the best-characterized pathway, CRM1-independent nuclear export mechanisms have been proposed for various proteins [5, 22, 23].
In conclusion, it is hard to define NES by its hydrophobic pattern only. For correct classification of leucine-rich regions with and without NES activity, detection of less apparent sequence features, secondary structure, flexibility and/or surface exposure is still necessary [19]. In addition, there are more complex signal sequences which exist, some of which confer both import and export functions [24]. In all cases, NLS and NES are constant constituents of the cargo and not removed during transport. They can be located at any site within a polypeptide and frequently occur in more than one copy per molecule [26].
Both BHV-1 VP8 and VP22 were identified to contain an NES and located in leucine-rich regions, consisting of amino acids 485LSAYLTLFVAL495 and 204LDRMLKSAAIRIL216 respectively. Moreover, the nuclear export activity of VP22 NES was completely blocked by LMB [36, 37]. Therefore, VP22 is a LMB-sensitive protein, which suggests that VP22 mediates nuclear export through a leucine-rich NES by a CRM-1-dependent pathway [37].
-
Many viral proteins shuttle between the cytoplasm and the nucleus to exert their specific biological functions. The transport of molecules across the nuclear membrane relies on the NPC, the transport carriers (including importins and exportins) and the Ran. Karyopherins bind to their cargoes by recognition of specific NLS or NES. Although transport mechanisms of molecules have been elucidated to a certain extent, many details remain unknown. With further investigation of the molecular basis of nuclear transport, it will significantly promote the design of anti-virus strategies and gene vector therapy.














 DownLoad:
DownLoad: