-
Dear Editor,
Rabbit hemorrhagic disease (RHD) is a highly contagious disease of both wild and domesticated rabbits (Oryctolagus cuniculus). The causative agent of the disease is the rabbit hemorrhagic disease virus (RHDV), belongs to the genus Lagovirus within the family Caliciviridae (Granzow et al., 1996; Ohlinger et al., 1990). It is a small and non-enveloped virus with a 7.5 kb single stranded positive sense RNA genome (Meyers et al., 1991; Meyers et al., 2000). Based on an analysis of VP60 sequences, six different gene clusters with one containing antigenic variants were identifed (Le Gall-Recule et al., 2003).
Recently, an RHDV variant with new genetic and epidemiological characteristics has been reported in China (Wang et al., 2012). To investigate the prevalence of RHDV, more convenient and sensitive diagnostic methods than hemagglutination assays (a widely used method for RHDV detection) are needed. SYBR based real-time PCR detection methods are commonly used in laboratories, since they do not require specifc reagents other than a pair of primers. In the present study, we developed an SYBR-based real-time PCR method and assessed RHDV loads in different body fluids and tissues of experimentally infected rabbits. In addition, clinical samples collected from diseased farm rabbits were tested with this method.
Two sets of primers targeting the conserved RHDV VP60 region were designed and used in a nested RTPCR, which served as a control. The external primers were VP60-D-Es (5′-GCAGTTCCGCTTCATA-3′) and VP60-D-Ea (5′-TGGTCAATGTCGGCAAAC-3′) for an amplicon of 540 bp. The internal primers were VP60-DIs (5′-GTTCCCACACTGGTCCTTAG-3′) and VP60-DIa (5′-GTGAGGACTGGGGTCGTGAG-3′) for an amplicon of 188 bp (nt 5863 to 6050 in the RHDV genome). The internal primers were also used in the real-time PCR assay. We optimized the PCR parameters and a mixture of 1.5 mmol/L MgCl2 and 1 µmol/L primers were used in the subsequent analyses. The sensitivity of the real-time PCR assay was determined by running serial ten-fold dilutions of a vector, pMD-18T-VP60, harboring the RHDV VP60 gene. The detection limit of the real-time PCR was approximately 6.09 plasmid copies/sample. The diluted pMD-18T-VP60 was also used to generate a standard curve. The standard curve of the threshold cycle (Ct) versus the log DNA concentration was linear within the range of 6.09 × 107 to 6.09 copies/µL, with a slope of –3.139 (R^2 = 0.997), suggesting an optimum PCR effciency of 2. This effciency corresponded to the amplification of one full-length target copy for every available template in each cycle. The melting peak analysis indicated that the melting temperature of the specifc amplicon was 83.7 ± 0.10 ℃.
The amplicons, corresponding to the 5′ terminus of VP60, were confrmed by sequencing. We also examined the specifcity of the proposed real-time PCR by testing other rabbit viruses, including rabbit myxomatosis virus (RMV), and bacteria, including Escherichia coli and Pasteurella multocida. As expected, no positive signal was recorded for any of the viruses or bacteria tested.
The sensitivities of conventional, nested, and real-time PCR for RHDV DNA were compared. The DNA concentration was measured spectrophotometrically and the copy number was calculated using an equation reported previously (Romanova et al., 2009). Serial dilutions were prepared, a total number of copies between 6.09 and 6.09 × 107 were amplifed, and the products were run in agarose gels (Figure 1A, 1B). Compared with the other methods, the dynamic range of conventional PCR was narrower, with a detection limit of 6.09 × 102 genomic copies of RHDV (Figure 1A). Nested PCR had a wider dynamic range and could detect 6.09 copies of DNA (Figure 1B). The real-time PCR method using SYBR green chemistry proposed in this work (with Ct < 35) had a detection range similar to that of nested PCR (Figure 1C). The amplifcation plot and melting curves demonstrated that the fluorescence observed was specific to RHDV DNA.
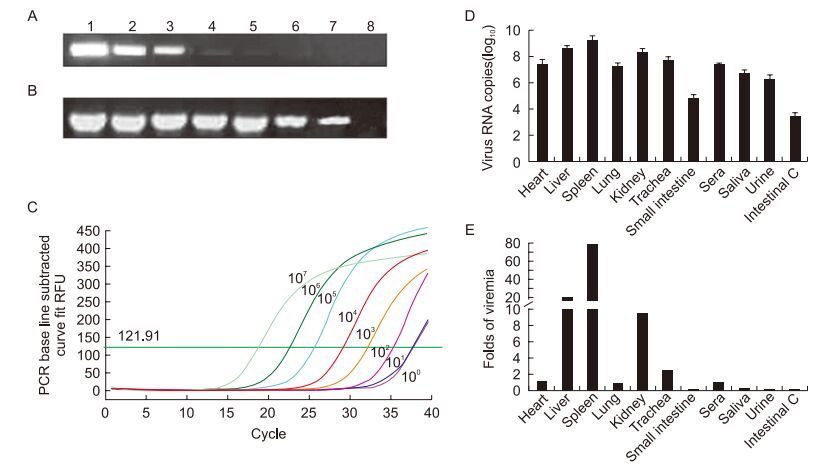
Figure 1. Analysis of the sensitivities of conventional PCR (A), nested PCR (B), and real-time PCR methods (C), and distribution of virus in experimentally infected rabbits (D, E). (A, B) Lanes 1–7: PCR results of serially diluted plasmids from 1.0 × 107 copies/μL to 1.0 × 101 copies/μL. Lane 8: negative control. (C) The amplifcation result of the real-time PCR with relative copies of the plasmid. (D) Average RHDV copy numbers in different samples (n = 3) and (E) a comparison of the RHDV load in different samples versus the RHDV load in sera (n = 3).
We further explored the use of this method in an anal-ysis of tissue distribution of the RHDV variant XA/2010/China in acute stage infection. Three rabbits infected with the isolate died within 72 h, and samples were collected immediately afterward. Negative samples were collected from uninfected rabbits euthanized by an overdose of sodi-um pentobarbital at 72 h. The viral genome load in different tissues, including the heart, liver, spleen, kidney, lung, trachea, and small intestine, and body fluids, including sera, urine, saliva, and intestinal contents, was determined by the real-time PCR method. Viral genome copy numbers in 100 mg (or 100 μL) of tissue are presented in Figure 1D. The highest number of viral RNA copies was found in the spleens, with an average value from three samples of 1.59 × 109. High numbers of viral RNA copies were also found in the livers and kidneys, with average viral copy numbers of 4.11 × 108 and 1.90 × 108, respectively. The number of viral copies in the heart, lung, trachea, sera, and urine were 2.17 × 107, 1.68 × 107, 4.99 × 107, 2.00 × 107, and 1.8 × 106, respectively. The lowest RHDV copy number was found in the small intestine, and particularly in the contents of the small intestine (Intestinal C) (5.67 × 104 and 2.51 × 103, respectively). RHDV virus copy numbers in the saliva were higher (5.01 × 106) than those in either urine or intestinal contents. All of the samples from the control group were negative (Ct ≥ 40), and all of the samples from the infected rabbits tested positive by realtime PCR (Ct < 35). To examine the distribution of the virus, the viral copies in tissues and liquid samples were compared with the virus load in the sera (Figure 1E). The virus replicates quickly in the spleen, liver, kidney, and trachea, and more viruses was observed in these tissues than in the sera. In the other organs, the virus may have been brought in by blood or tissue fluid, because the level of virus was lower in these organs than in the serum.
Real-time PCR methods have also been used for the detection of RHDV in clinical samples. The spleens and/ or liver samples from 83 rabbits were collected from 9 RHDV vaccine-using rabbit farms in the Shaanxi or Henan provinces. A total of 47 samples were positive with Ct values of less than 35 (Table 1). Another 10 samples were suspected positives because they had Ct values greater than 35, but less than 40. The other 26 samples were negative, as the Ct values were greater than 40 without a resulting melting curve. In almost all positive cases, we detected a low level of infection with immunity against RHDV. Most Ct values (44/47) of the positive samples were around 30, and all the rabbits were from farms where the vaccine had been introduced, so the rabbits should have gained immunity from either vaccination or maternal antibody. Nevertheless, the high level of RHDV in the diseased rabbits was a surprise. Based on our survey, in most farms with immunized rabbits, RHDV is circulating with sub-acute or chronic disease. RHDV infections with concurrent immunity against the virus have also been reported in wild rabbits (Capucci et al., 1998; Strive et al., 2009). This may be caused by variation in the viral load or a low level of immune response.

Table 1. Detection of rabbit hemorrhagic disease virus in clinical samples.
In this study, we developed a real-time PCR assay for RHDV detection and quantitation. With this method, the distribution of RHDV in the internal organs and body fluids of infected rabbits was analyzed. The highest viral RNA load was found in the spleen, followed by that in the liver. The virus is shed mainly through the oral, nasal, and urethral routes. Through testing of clinical samples, we found that most farmed rabbits that had been immunized had low levels of RHDV in their tissues. These fndings may be useful for further research on the pathogenic mechanism of RHDV and potential RHDV receptors in organs.
HTML
-
This research was supported by the Shaanxi Agriculture Science and Technology Research Projects (2014K02-06-01). We acknowledge Dr Yi Li (Central China Normal University) for providing the RMV virus DNA sample used in specifcity test. The authors declare that they have no competing interests. All the animal tests comply with Shaanxi province laboratory animal management approach and the requirement of animal welfare.















 DownLoad:
DownLoad: