HTML
-
Coronaviruses(CoVs)are classified into four genera, namely alpha-, beta-, gamma-and deltacoronavirus, under the family of Coronaviridae and the order of Nidovirales(Woo et al., 2012). The first three genera were previously known as groups I, II and III, respectively(Lau et al., 2006; Zhong et al., 2012). CoVs have been shown to infect many different hosts including bats, birds, dogs, mice and human(Woo et al., 2009; de Groot et al., 2013). The infections are commonly zoonotic in nature(Chan et al., 2013). In the past 50 years, several human CoVs(HCoVs)were identified. HCoV-229E and HCoV-OC43, belonging to alpha- and betacoronavirus respectively, were the first two HCoVs identified in the mid-1960s(Tyrrell and Bynoe, 1965; Hamre and Procknow, 1966; Mclntosh et al., 1967). Healthy individuals infected with either HCoV-OC43 or HCoV-229E develop illnesses within the range of typical common colds with good prognosis(Bradburne et al., 1967). Since the identification of these two HCoVs, extensive studies were conducted to understand their pathogenicity. However, almost all studies showed that HCoV-OC43 and HCoV-229E caused mild illnesses with high titers of neutralizing antibodies(Bradburne et al., 1967). The idea of HCoV being a relatively weak respiratory disease-causing agent was therefore presented to the field.
This idea was generally accepted until the outbreak of SARS in 2003. SARS-CoV was the first HCoV identified to cause acute respiratory distress syndrome(ARDS)(Cheng et al., 2007; Graham et al., 2013). According to World Health Organization(WHO), a total of 8096 cases from 29 countries were reported with a case mortality rate of 9.6%. The SARS outbreak changed the landscape of CoV studies entirely and marked the new era of combating infectious diseases. Tremendous efforts have been put into understand ing SARS-CoV pathogenicity, opening a new page of CoV biology. Despite advances in infection control and quarantine measures in the past decade, another HCoV causing ARDS was identified in Saudi Arabia as a novel lineage C betacoronavirus in September 2012(Zaki et al., 2012). The newly identified HCoV was later named MERS-CoV. Up to October 2015, 1611 laboratory-confirmed cases were reported to WHO with 575 related deaths in 26 countries, including a recent outbreak involving 186 cases and 37 deaths in South Korea. MERS-CoV is closely related phylogenetically to two bat CoVs, HKU4 and HKU5, shedding light on the possible zoonotic reservoir of MERS-CoV(Zaki et al., 2012; Memish et al., 2013).
Together with HCoV-HKU1 identified in 2005(Woo et al., 2005) and HCoV-NL63 discovered in 2004(Fouchier et al., 2004; van der Hoek et al., 2004), six HCoVs have been documented up to date. These 6 HCoVs present diseases with a range of clinical severity from typical common cold in HCoV-OC43, HCoV-229E, HCoV-HKU1 and HCoV-NL63 to ARDS in SARS-CoV and MERS-CoV. Why these CoVs show dramatically different pathogenicity in human is an important but unanswered question in the field. One model to explain this difference is based on adaptation and host immunity. According to this model, bats are reservoir of various CoVs. Bat CoVs constantly emerge in human via intermediate hosts such as civets and dromedaries. Exposure of immunologically na ve human populations to these CoVs commonly causes severe diseases plausibly due to aberrant activation of innate immunity and lack of immune memory. When some CoVs become better adapted in human by acquiring the ability to transmit from human to human readily, pandemics could arise. Meanwhile, as they become fully adapted, the CoVs might only cause mild diseases in human. Existing evidence supports the origin of HCoV-OC43, HCoV-229E, HCoV-HKU1 and HCoV-NL63 from bats and other animals(Woo et al., 2009; Huynh et al., 2012; Corman et al., 2015). Adaptation and virus-host interaction are also known to be major determinants in CoV pathogenesis(Pepin et al., 2010; Chan et al., 2013). It will therefore be of great interest to see whether emerging human CoVs might be particularly capable of evading innate antiviral response while activating pathological inflammation. In other words, we need to determine whether the more severe clinical presentations might be accounted for by the specific interaction between host and emerging human CoVs, namely SARS-CoV and MERS-CoV. In this review, the host innate antiviral response to CoV infection is particularly focused. In addition, the viral strategies adopted by SARS-CoV and MERS-CoV to subvert innate immunity are also summarized to provide inspiring insights that may explain the discrepancies in virulence(Figure 1).
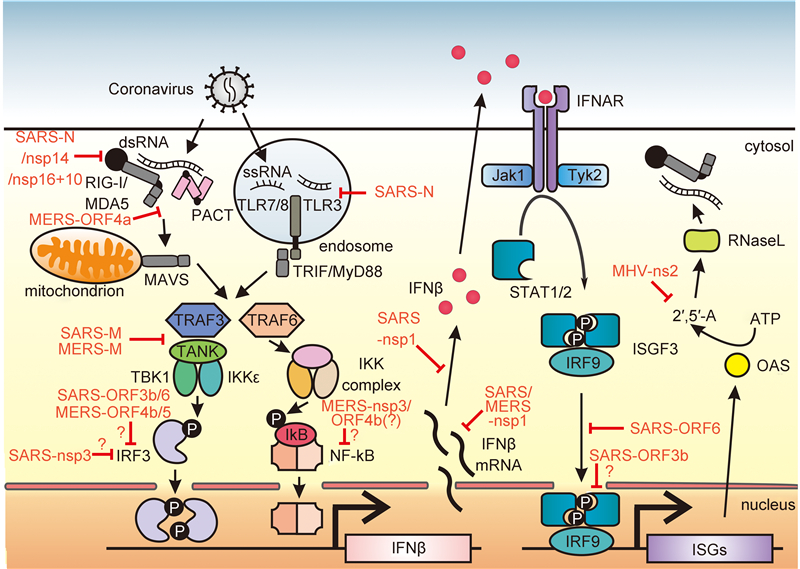
Figure 1. Innate immune response mediated against coronavirus infection and viral evasion mechanisms. (Left) Upon CoV infection, viral genome ssRNA as well as dsRNA intermediate found in virus life cycle are exposed to host innate immune sensors, RIG-I/MDA5 in cytoplasm or Toll-like receptors TLR3/7/8 in endosome. Activation of these immune sensors initiates a downstream signaling cascade that leads to IFN-β gene expression. RIG-I/MDA5 conveys signal through a mitochondrial adaptor MAVS while TLR signals through TRIF/MyD88. Both pathways share the common TRAF adaptor to activate transcription factors. TRAF3 serves as an adaptor which activates TANK × TBK1/IKKε complex for IRF3 phosphorylation and subsequent dimerization, while TRAF6 is responsible for the activation of IKK complex which phosphorylates the canonical inhibitor of NF-κB (IκB). Activated transcription factors are translocated into the nucleus to drive IFN-β expression. (Right) IFN-β are secreted into extracellular space and bound to its cognate receptors IFNAR to activate downstream JAK-STAT signaling. Receptor-associated tyrosine kinases Jak1 and Tyk2 are brought to juxtaposition for self-phosphorylation and activation. STATs are recruited to and phosphorylated by the tyrosine kinases. Phosphorylated STAT1/2 with IRF9 forms a ternary complex ISGF3 which translocates into the nucleus and binds to ISRE in the promoter region upstream of ISG genes. ISG genes are expressed consequently to establish an antiviral state in cells. OAS is an example of ISG which produces 2′, 5′-oligoadenylate (2′, 5′-A) upon detection of dsRNA and activates RNase L to cleave viral RNA to yield more RLR ligand as a positive-feedback mechanism of IFN production. The CoV-encoded proteins shown in red are known to intervene the host innate immune signaling at various action points as evasion mechanisms to sustain viral replication and propagation. The action points at which viral proteins function marked with a question mark (?) represent controversial and inconclusive findings in the field or molecular mechanisms not well studied. MHV: mouse hepatitis virus.
-
CoVs are polycistronic positive-sense single-stranded RNA(ssRNA)viruses with genomes of about 30 kb in size. The 5′ most two-thirds of CoV genome encodes polyprotein 1a(pp1a) and pp1ab replicase polyproteins, which are further cleaved by viral proteases to yield non-structural proteins(nsps), while the 3′ end of the genome encodes structural and lineage-specific proteins(Durai et al., 2015). The CoV life cycle begins with the binding to cellular receptor followed by membrane fusion as well as viral RNA and protein synthesis in the cytoplasm. The pp1a and pp1ab polyproteins are co-translationally processed resulting in the formation of the replicase complex. A set of nested subgenomic mRNAs and genomic RNA, which possess both the same 3′ end and a common 5′ leader sequence derived from the 5′ end of the genome, is then transcribed. Normally, only the 5′ end of each mRNA is translated. Virion assembly is achieved by budding into intracellular membranes and virion release is accomplished through the secretory pathway(Cheng et al., 2007; Durai et al., 2015).
The coronaviral spike(S)protein is responsible for binding to specific host receptor on cell surface and fusing viral envelope with lipid membrane of host upon infection(Bosch et al., 2003; Rota et al., 2003; Chen et al., 2013). HCoV-NL63 and SARS-CoV from α-and β-genera respectively recognize angiotensin-converting enzyme 2(ACE2)(Li et al., 2003; Pyrc et al., 2007; Frieman et al., 2008; Chen et al., 2013)while MERS-CoV infects cells through another cell surface enzyme dipetidyl peptidase 4(DPP4)(Chen et al., 2013; Raj et al., 2013). Aminopeptidase N(APN)has also been found to be recognized by some α-genus CoVs like HCoV-229E(Yeager et al., 1992). Cell surface receptor binding dictates species-specific viral entry as well as tropism. This also confines the direction of cellular antiviral response. We and others have shown the ability of CoV S proteins to activate unfolded protein response and endoplasmic reticulum stress(Chan et al., 2006; Fung et al., 2014; Siu et al., 2014b). The activity of S might also be functionally related to coronaviral perturbation of innate antiviral response including IFN and cytokine production.
-
Pattern recognition receptors(PRRs)constitute an indispensable part of the host innate immune defense mechanism by the detection of foreign, non-self patterns from invading microbes distinct from host. These pathogen-associated molecular patterns(PAMPs)are usually biomolecules derived from the surface or generated during the life cycle of the microbes. The detection of PAMPs by host PRRs activates innate immune response including the expression of type I IFNs and cytokines for clearance of invading microbes. During CoV infection, retinoic acid-inducible gene I(RIG-I)-like receptors(RLRs) and Toll-like receptors(TLRs)are believed to bear pivotal importance in stimulating host type I IFN induction. It is therefore essential to review the sensing mechanism of the PRRs to understand viral evasion mechanisms and provide insights on the development of potential viral antagonists.
-
After viral entry, CoV genomes are exposed in the cytoplasm for expression of viral proteins, providing an opportunity for viral RNA sensing by host. RLRs are ubiquitously expressed cytoplasmic RNA helicases of DExD/H box family responsible for sensing double-stranded RNA(dsRNA)(Yoneyama et al., 2005). Three types of RLRs have been identified up to now, including RIG-I, melanoma differentiation-associated gene 5(MDA5) and laboratory of genetics and physiology 2(LGP2)(Loo and Gale, 2011). RIG-I and MDA5 consist of N-terminal caspase activation and recruitment domain(CARD)in two tandem copies, a central DExD/H box helicase domain and a C-terminal domain(CTD)(Yoneyama et al., 2004, 2005). The N-terminal CARDs are the effector domain of RLRs to mediate downstream transduction, which is held by the CTD when unstimulated(Jiang et al., 2011; Kowalinski et al., 2011; Luo et al., 2011). However, in the presence of residual amount of cytoplasmic dsRNA, RLRs bind to dsRNA through the central DExD/H box helicase domain and CTD with ATP, causing a conformational change that exposes the N-terminal CARDs for signal transduction(Yoneyama et al., 2004; Jiang et al., 2011). LGP2 lacking the N-terminal CARDs is thought to act as co-factor that augments the function of RIG-I and MDA5(Satoh et al., 2010; Bruns et al., 2014). Exposure of CARDs leads to oligomerization of RIG-I or MDA5 to form filamentous structure(Berke et al., 2012; Peisley et al., 2013; Wu et al., 2013). The CARD filament recruits and further initiates similar filamentous structure formation of CARD on MAVS, an adaptor protein which further recruits downstream effectors tumor necrosis factor receptor-associated factor 3(TRAF3), TANK-binding kinase 1(TBK1) and IκB kinase ε(IKKε)(Loo and Gale, 2011; Wu et al., 2014). TBK1 and IKKε form a complex of activated protein kinase for phosphorylation and activation of not only MAVS adaptor(Liu et al., 2015a), but also IRF3 transcription factor(Loo and Gale, 2011). Activated IRF3 are phosphorylated, dimerized and eventually translocated to the nucleus. On the other hand, TRAF2/6 is also recruited to MAVS for NF-κB activation. Specifically, canonical NF-κB inhibitor IκB is phosphorylated and then degraded through proteasomes in a ubiquitination-dependent fashion(Loo and Gale, 2011). IκB degradation exposes nuclear localization signal on NF-κB dimer for nuclear translocation. Activated IRF3 and NF-κB together with other transcription factors including c-Jun assemble the enhanceosome that binds to IFN-β promoter for IFN-β expression(Ford et al., 2010; Loo and Gale, 2011). Infection with mouse hepatitis virus induces RIG-I expression. In addition, the activation of type I IFN production by this CoV in oligodendrocytes requires both RIG-I and MDA5(Li et al., 2010). Thus, RLRs might play an important role in the sensing of CoV infection.
Several critical questions concerning RLR recognition of CoVs merit further investigations. First, the role of RLRs in CoV sensing should be studied in RLR-null and CoV-susceptible cells and animals. When necessary CRISPR/Cas9 technology might be used to disrupt RLR genes in target cells(Hsu et al., 2014; Yuen et al., 2015). Second, the CoV PAMPs recognized by RLRs should be identified and characterized. Particularly, it will be of interest to see whether and how common and highly structured regions in coronaviral genome, such as the aforementioned 5′ leader sequence, might be recognized by RLRs. For example, a polyuridine motif in the 3′ untranslated region of hepatitis C virus genome and the panhandle structure in RNA viruses such as influenza A virus have previously been shown to be RIG-I agonists(Saito et al., 2008; Weber et al., 2013; Kell et al., 2015; Liu et al., 2015b). In addition, possible involvement of viral proteins such as nucleocapsid(N)in this recognition as in the case of other RNA viruses(Saito et al., 2008; Weber et al., 2013)should also be clarified. Finally, comparative analysis of SARS-CoV, MERS-CoV and other HCoVs for their ability to activate RLRs will shed light on whether RLR activation would be a critical determinant in CoV virulence.
-
CoVs have been observed to infect host cells through more than one pathway. While CoV entry by the fusion of viral envelope and host membrane has been described, the endosomal pathway is still considered the classical entry pathway for CoVs. In this pathway the activation of S protein cleavage by cathepsin L and transmembrane serine protease TMPRSS2 occurs in the absence of cell surface proteases in certain cell types(Shirato et al., 2013; Burkard et al., 2014). In this regard, TLR family may play an essential role in sensing CoV infection through the endosomal pathway. TLR family was identified as another PRR homologous to Drosophila Toll receptor(Boehme and Compton, 2004), sensing various PAMPs within the endosome which leads to induction of cytokines and IFNs. In human, each of the 11 TLRs is known to specifically recognize a particular PAMP and preferentially resides in either plasma or endosomal membrane. The cellular localization of TLRs defines their functions in detecting different PAMPs. For example, TLRs critically involved in viral nucleic acid sensing, including TLR3 for dsRNA, TLR7 and TLR8 for ssRNA, and TLR9 for unmethylated CpG island of dsDNA viruses, are mainly localized in endosomal membrane while other members having a role in sensing other biomolecules derived from microbial surface components localized to plasma membrane of infected cells(Xagorari and Chlichlia, 2008; Kawai and Akira, 2010). TLR family members being type 1 transmembrane proteins share a similar structure with a single transmembrane domain. TLR specificity is determined by the ectodomain made up of various number of leucine-rich repeats(LRRs)that bind the corresponding PAMP directly(Boehme and Compton, 2004). Signal transduction begins with ligand binding to LRRs in the ectodomain, thus recruiting cytosolic adaptor protein MyD88 with cytoplasmic Toll/IL-1 receptor(TIR)domain by homotypic TIR-TIR domain interaction(Xagorari and Chlichlia, 2008). The TLR-MyD88 complex then recruits and activates interleukin 1R-associated kinase(IRAK)by phosphorylation. The activated IRAK then in turn associates with TRAF6 and activates a series of downstream effectors leading to the activation of a range of cytokines and IFN-stimulated genes(ISGs), while activation of type I IFN expression by TLR3 is independent of MyD88 but dependent on TRIF(Boehme and Compton, 2004; Xagorari and Chlichlia, 2008). TLR pathway is significantly involved in the suppression of CoV replication and induction of type I IFN expression. Mice deficient of either TLR3 or TLR4 were more prone to SARS-CoV pathogenesis(Mazaleuskaya et al., 2012; Totura et al., 2015). Notably, disruption of either MyD88 or TRIF arm of the TLR signaling pathway causes lethal SARS-CoV disease, indicating the importance of both arms in host innate immunity against SARS-CoV(Totura et al., 2015). Full characterization of the role of TLRs in host innate antiviral response against SARS-CoV and MERS-CoV versus other HCoVs will not only provide new knowledge about how TLR activation might impact CoV pathogenesis, but might also identify new strategies for antiviral and vaccine development. For example, synthetic TLR agonists could potentially serve as antivirals and vaccine adjuvants in the prevention and control of CoVs.
RIG-I-like receptors
Toll-like receptors
-
Innate antiviral response is the first line of defense against CoV infection. Type I IFNs are important antiviral and immunomodulatory agents. Type I IFNs function by binding to IFN-α receptor-1(IFNAR-1) and IFNAR-2 receptor complex, thus activating Janus family tyrosine kinase(JAK), leading to the phosphorylation of signal transducer and activator of transcription(STAT), a family of transcription factors regulating the expression of ISGs. Activated STAT and IRF9 form IFN-stimulated gene factor 3(ISGF3), stimulating expression of ISGs by binding to IFN-stimulated response element(ISRE)in promoters of ISGs(Levy et al., 2001; Samuel, 2001). Viral induction of ISGs was abrogated in STAT1-/- mice infected with SARS-CoV. The viral infection could not be cleared resulting in severe disease, extensive lung injury and 100% mortality(Frieman et al., 2010; Zornetzer et al., 2010). This indicates the importance of STAT1 in SARS-CoV pathogenesis.
ISGs are the workhorses of the innate antiviral response with diverse functions including direct antiviral activities and regulation of adaptive immune system(Schneider et al., 2014). For example, IFN-inducible gene p53 evokes apoptosis in virus-infected cells(Takaoka et al., 2003). IFN-inducible protein kinase PKR, 2′, 5′-oligoadenylate synthetase(OAS) and RNase L are important modulators involved in dsRNA sensing, viral gene expression and replication. They act sequentially to trigger viral RNA degradation and suppression of viral activities(Samuel, 2001). Other ISGs encoding antiviral effectors such as Mx proteins, cholesterol-25-hydrolse, IFITM proteins, TRIM proteins, viperin, tetherin, cGAMP synthase and STING could also be highly relevant to CoV infection(Schneider et al., 2014; Schoggins et al., 2014; Ma et al., 2015a; Ma et al., 2015b). Inflammatory responses triggered by inflammatory cytokines like tumor necrosis factor α(TNF-α) and IFN-γ are also found to be IFN-dependent(Samuel, 2001). IFNs do not only exert antiviral effects through activation of innate immunity but also act as modulators of adaptive immunity. Adaptive immune response is activated by increased level of IFNs. The levels of major histocompatibility complex(MHC)proteins class I and II are found up-regulated by IFNs. This facilitates efficient antigen presentation and hence cellular immune response to CoV infection(Samuel, 1991, 2001; Ivashkiv and Donlin, 2014). In addition, the roles of non-conventional ISGs including microRNAs, long non-coding RNAs and alternatively spliced isoforms have been increasingly recognized in recent years(Schneider et al., 2014). It will be of importance to determine whether SARS-CoV and MERS-CoV might be unique in ISG activation as suggested in a recent study, which demonstrated that MERS-CoV induces repressive histone modifications to down-regulate specific subsets of ISGs(Menanchery et al., 2014b). In relation to this, two areas concerning ISG activation by CoVs might require more attention and research efforts. First, unbiased and large-scale screening of antiviral ISGs using RNA interference or CRISPR/Cas9 technology might be carried out to identify key cellular factors that restrict SARS-CoV and MERS-CoV replication and infection. Second, small-molecule compounds that activate antiviral ISGs could be identified and tested for inhibition of SARS-CoV and MERS-CoV replication and infection. For example, establishing the significance of cGAS and STING in CoV infection might lead to the development of cyclic dinucleotides such as c-di-GMP and cGAMP as novel anti-CoV agents.
-
CoVs have been reported to directly or indirectly suppress IFN production and signaling pathways by a subset of viral proteins via various mechanisms. In many cases, infected patients have shown diminished levels of type I IFNs. This is especially true for SARS and MERS patients with severe diseases(Faure et al., 2014). It was also shown that SARS-CoV and MERS-CoV were capable of evading type I IFN production and signaling to different extents in cultured cells(Kindler et al., 2013). When the deficiency in type I IFN production in CoV-infected cells was remedied by IFN-α treatment, CoV replication was inhibited(Falzarano et al., 2013). Combination of IFN-α with other antiviral drugs further improves the survival of infected patients(Omrani et al., 2014). This evidence suggests an essential role of type I IFNs in the antiviral effect against CoV infection. CoVs have evolved strategies to counter host antiviral response by antagonizing type I IFN production and signaling. CoV proteins have been characterized to exhibit innate immunosuppressive effects in cellular models. Below we will discuss them in three categories: structural, lineage-specific and non-structural proteins(nsps)(de Groot et al., 2013). Nsps of CoVs are involved in the assembly of the replicase complex for viral RNA synthesis(Sevajol et al., 2014). Certain nsps have also been reported to possess innate immunosuppressive effect that facilitates viral replication and propagation, although these proteins per se are not required for viral life cycle(Narayanan et al., 2008b; Lokugamage et al., 2015). Nsps of different CoVs are more or less evolutionarily conserved suggesting their functional significance, with the exception of nsp1 and nsp2, which are thought to contribute to virulence of certain CoVs(Neuman et al., 2014). Four structural proteins are found in CoVs, namely S, membrane(M), envelope(E) and N proteins. Structural proteins contribute the architecture for virion assembly. Accessory proteins are lineage-specific with diverse behaviors in different CoVs but are not essential for viral replication and propagation(de Groot et al., 2013).
-
CoV nsps have shown suppressive effects in various immune pathways including type I IFN production and signaling. SARS-CoV and MERS-CoV nsp1 proteins have been shown to selectively induce degradation of host mRNA by inducing endonucleolytic cleavage while leaving viral RNAs intact(Huang et al., 2011; Lokugamage et al., 2015). In addition to the induction of endonucleolytic cleavage of host mRNA, general inhibition of host mRNA translation is achieved by binding of 40S subunit of ribosome with SARS-CoV nsp1(Huang et al., 2011). Particularly, SARS-CoV nsp1 inhibits innate immune response by translational repression of IFN mRNA transcripts, hence altering IFN production and signaling(Narayanan et al., 2008a; Tanaka et al., 2012). MERS-CoV nsp1 has also been characterized to specifically induce endonucleolytic cleavage of nuclear transcribed mRNA while sparing cytoplasmic host mRNA and viral RNA(Lokugamage et al., 2015). This suggests a novel mechanism for evading host immune response.
CoV nsp3 protein has been characterized with a papain-like protease(PLpro)domain for enzymatic cleavage of pp1a and pp1ab as well as a PLP2 domain with deubiquitinating and deISGylating activity(Clementz et al., 2010; Mielech et al., 2014). MERS-CoV PLpro is able to antagonize IFN production induced by RIG-I and MDA5 as well as NF-κB activation(Mielech et al., 2014). MERS-CoV PLpro is catalytically more efficient(Báez-Santos et al., 2014) and its catalytic activity is indispensable for the suppressive effect on RIG-I, MDA5 and NF-κB(Mielesh et al., 2014). In contrast, SARS-CoV PLpro does not require enzymatic activity for IFN antagonism(Clementz et al., 2010). HCoV-NL63 and SARS-CoV PLP2 transmembrane domain can also act as potent IFN antagonists to suppress IFN production induced by RIG-IN, a dominant active form of RIG-I(Clementz et al., 2010). In another view of direct inhibition of IFN induction, nsp3 with deubiquitinating and deISGylating activity may also influence the ubiquitination and ISGylation pattern and dynamics thus indirectly hindering innate immune response against CoV infections(Clementz et al., 2010). For example, ISGylation and ubiquitination of IRF3 required for optimal activation is probably altered by PLP domain of nsp3.
Apart from directly manipulating the signaling pathway involved in IFN production, several CoV nsps were identified to act on viral RNA to minimize IFN stimulation. N7-methylguanosine is the fundamental moiety of eukaryotic mRNA cap structure and 2′-O-methylation on this moiety is a representative host signature to avoid PRR activation as well as ISG action. Particularly, viral RNA with this modification evades recognition by MDA5 or IFIT family antiviral factors(Züst et al., 2011; Daffis et al., 2010). This is a common immunoevasive mechanism adopted by not only different CoVs but also other RNA viruses. Functional screening in yeasts suggested a novel function of SARS-CoV nsp14 as a guanine-N7-methyltransferase, the activity of which is required for viral replication and transcription(Chen et al., 2009). Another nsp of SARS-CoV, nsp16, also possesses 2′-O-methyltransferase activity(Menachery et al., 2014a; Menachery et al., 2014c). Structural modeling suggested that SARS-CoV nsp16 associates with nsp10 in 1:1 ratio to form a complex of mature 2′-O-methyltransferase for viral cap methylation(Chen et al., 2011; Decroly et al., 2011). A short peptide derived from nsp10 conserved region has been shown to be a promising nsp16 antagonist which outcompetes native nsp10 to blunt 2′-O-methyltransferase activity and restrict viral replication(Wang et al., 2015). Plausibly, CoV nsps might execute their innate immunosuppressive roles by targeting type I IFN production and signaling. Further investigations are required to clarify whether and how far the sensing of CoV RNA and the induction of innate antiviral response are involved in the inhibitory activity of the nsp antagonists on CoV replication.
-
CoV structural proteins have been shown to inhibit IFN production and signaling at multiple levels. SARS-CoV N protein showed inhibitory effects on IFN production induced by Sendai virus and dsRNA analogue poly(I:C)but no inhibition could be observed when downstream signaling molecules of TLR and RLR pathway were overexpressed. Truncation mutant of N protein shows that the C-terminal domain is critical for RNA-binding and IFN-antagonizing effect(Lu et al., 2011). This suggests SARS-CoV N may interfere with RNA recognition by host immune sensors such as RIG-I and MDA5 thus achieving suppressive role in IFN production. Other than N protein, SARS-CoV M protein has been characterized to potently down-regulate IFN production by impeding the formation of TRAF3·TANK·TBK1/IKKε complex through the first transmembrane domain(Siu et al., 2009, 2014a). SARS-CoV M protein inhibits IFN production possibly through a sequestration model in which components of TRAF3·TANK·TBK1/IKKε complex, an active complex for IRF3 phosphorylation, are sequestered to specific locations in the cell(Siu et al., 2009). SARS-CoV M protein therefore exerts its inhibitory effects by impeding the formation of TRAF3·TANK·TBK1/IKKε complex but not by modulating the catalytic activity of the complex.
MERS-CoV M protein also exhibits IFN-antagonizing effects similar to its counterpart in SARS-CoV. In a previous study, MERS-CoV M is shown to impede IFN production by preventing IRF3 translocation into the nucleus(Yang et al., 2013). However, the detailed mechanism of inhibition remains unknown. Recently, our group has characterized the mode of inhibition of IFN production by MERS-CoV M. Consistently with previous report, we show that MERS-CoV M suppresses IFN production by preventing IRF3 activation. We showed that MERS-CoV M interacts with TRAF3 which impedes the recruitment of TBK1 to TRAF3 complex. IRF3 activation and dimerization have also been hampered as a result. The inhibitory effect is at least in part accounted for by the N-terminal transmembrane domains. Despite of the similar behaviors, MERS-CoV M can only moderately suppress IFN expression when compared to SARS-CoV M. Interestingly, HCoV-HKU1 M protein does not exert any inhibitory effects on IFN production(Siu et al., 2014a), suggesting that the IFN-antagonizing activity of structural proteins is unique to each CoV but not universal. It will be of great interest to see whether this may correlate with the pathogenicity of different HCoVs.
-
Eight accessory proteins have been identified in SARS-CoV and five are found in MERS-CoV(Narayanan et al., 2008b). SARS-CoV genome encodes ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8a, ORF8b and ORF9b as accessory proteins(Narayanan et al., 2008b). SARS-CoV ORF3b and ORF6 have been found to antagonize type I IFN production and signaling. Particularly, SARS-CoV ORF3b and ORF6 suppress IFN-β production by perturbing IRF3 activation induced by Sendai virus infection. SARS-CoV ORF3b and ORF6 also suppress IFN-β-induced activation of ISRE in ISG promoters(Kopecky-Bromberg et al., 2007), although they are not able to reduce the level of phosphorylation of STAT1, a transcription factor that activates ISRE activity once phosphorylated. However, SARS-CoV ORF6 has been shown to inhibit STAT1 translocation for ISRE activation(Kopecky-Bromberg et al., 2007). The findings suggest a mode of inhibition of IFN-β signaling by SARS-CoV.
IFN antagonism of accessory proteins has also been observed in another deadly HCoV. MERS-CoV genome encodes ORF3, ORF4a, ORF4b, ORF5 and ORF8b(de Groot et al., 2013). Among the five accessory proteins, ORF4a, ORF4b and ORF5 show the ability to dampen IFN production(Yang et al., 2013). Suppression of IFN-β promoter-driven luciferase activity has been observed in cells transfected with ORF4a, ORF4b and ORF5 plasmids. All these 3 accessory proteins are able to block IRF3 translocation to the nucleus to activate IFN promoter(Yang et al., 2013). MERS-CoV ORF4a shows an additional level of inhibition of innate immunity by intervening NF-κB activation. In another study, ORF4a has been shown as an antagonist of IFN production by inhibiting IRF3 translocation but has no effect on IFN signaling(Niemeyer et al., 2013). Our group demonstrated that MERS-CoV ORF4a interacts with PACT, a cellular dsRNA-binding protein that optimally activates RIG-I-and MDA5-induced type I IFN production, in an RNA-dependent manner(Siu et al., 2014c). This suggests that ORF4a may compete with RIG-I and MDA5 for RNA, rendering the inactivation of RIG-I and MDA5. Direct interaction of ORF4a with PACT may also prevent interaction of PACT with RIG-I and MDA5, thus compromising PACT-dependent activation of RIG-I and MDA5 required for optimal induction of IFN production. Although we and others have observed the IFN-antagonizing activity of MERS-CoV ORF4b, different activity profiles and mechanisms have been suggested(Yang et al., 2013; Matthews et al., 2014). One recent report suggested that ORF4b directly interacts with and inhibits TBK1/IKKe in the cytoplasm but might also perturb type I IFN production in the nucleus through an unknown mechanism(Yang et al., 2015).
Mouse hepatitis virus, another betacoronavirus closely related to HCoV-OC43 and HCoV-HKU1, encodes a lineage-specific accessory protein named ns2 with innate immunosuppressive property(Zhao et al., 2012). Biochemical assays indicate that ns2 protein has phosphodiesterase activity against 2′, 5′-A, the product of OAS(Zhang et al., 2013). Thus, ns2 is a potent inhibitor of an IFN effector molecule and it might represent a new family of viral and cellular proteins with innate immunosuppressive activity(Zhang et al., 2013; Gusho et al., 2014). Whether distantly related proteins in HCoV-OC43 and HCoV-HKU1 might have similar activity remains to be determined. More importantly, it will be of interest to see whether SARS-CoV and MERS-CoV might encode proteins with similar enzymatic activity.
Multiple IFN antagonists have been identified and characterized in SARS-CoV and MERS-CoV. Some differences between these IFN-antagonizing viral proteins and their counterparts in other CoVs such as the parental bat viruses of MERS-CoV have also been noticed(Siu et al., 2014c). Existing evidence supports several important notions. First, although SARS-CoV and MERS-CoV share some features in common, they are distinct and use unique mechanisms for innate immune evasion(Perlman and Zhao, 2013). Second, both SARS-CoV and MERS-CoV are bat-origin CoVs that are well adapted in bats but newly emerge in human. This provides a golden opportunity for the study of CoV-host interaction, CoV adaptation as well as the arms race between host innate antiviral immunity and CoVs. Observing how the arms race between the host and SARS-CoV or MERS-CoV might evolve when the viruses become adapted to human will be most revealing and could provide important clues as to how a balance of power in this arms race might result in attenuation with increased transmissibility. Finally, studies on SARS-CoV and MERS-CoV have overturned existing concepts and derived new principles and thoughts to CoV biology. Particularly, mechanisms by which SARS-CoV and MERS-CoV evade innate immunity have attracted increasing attention. However, many key issues remain obscure. Particularly, better in vivo evidence should be obtained to clarify whether more potent inhibition of innate IFN production and signaling by SARS-CoV and MERS-CoV is a key determinant in virulence and disease severity.
Non-structural proteins
Structural proteins
Accessory proteins
-
CoVs have drawn a lot of interests in the light of the recent emergence of MERS-CoV. It remains to be understood whether the emerging deadly CoVs causing ARDS might ultimately be established and adapted in human resulting in significant attenuation of virulence. From the identific-ation of the first two HCoVs, HCoV-229E and HCoV-OC43 in the mid-1960s, we learned that HCoV was able to cause only common cold. However, the outbreaks of SARS and MERS that have claimed hundreds of lives revealed the other extreme of CoV pathogenicity and raised new questions in CoV biology. So far no vaccines have been developed against SARS-CoV and MERS-CoV.
Infection with SARS-CoV and MERS-CoV has been accompanied with suppression of innate immune response, most notably with the suppression of type I IFN production and signaling pathways. As the first-line defense in the immune system, suppression of innate immune response by these CoVs has impeded the host ability to restrict infection, causing significant casualties. Although many reports have shed light on the molecular mechanism by which various CoV proteins antagonize type I IFN production and signaling, most of the studies were performed with overexpression experiments in cellular models. Future emphasis should be put on the characterization of knock-out viruses with which the function of a particular viral gene could be studied in a more physiologically relevant context. Infectious clones and replicons for SARS-CoV and MERS-CoV have been generated for this reverse genetic approach(Yount et al., 2003; Almazán et al., 2006, 2013, 2014; Scobey et al., 2013). IFN and cytokine profiles of deadly HCoVs such as SARS-CoV and MERS-CoV can be compared with HCoV-229E and HCoV-OC43 causing mild diseases. The pivotal significance of type I IFNs in innate immune activation and modulation has been discussed in this review. Suppression pattern of IFN may provide insights on the high pathogenicity of deadly HCoVs. The arms race between host innate antiviral response and emerging human CoVs might evolve after their introduction and establishment in human populations, with significant impact on virulence, transmissibility and disease severity. Emerging human CoVs remain a potential threat to global public health. New knowledge about the host-CoV arms race will provide new ideas, targets and attenuated strains for the design and development of antivirals and vaccines for prevention and control of deadly CoV infections.
-
We thank Hinson Cheung, Kitty Fung, Edwin Kong and Sam Yuen for reading the manuscript critically. Coronavirus research in our laboratory was supported by Hong Kong Health and Medical Research Fund(13121032, 14130822 and HKM-15-M01) and Hong Kong Research Grants Council(HKU1/CRF/11G, C7011-15R and T11-707/15-R).
-
The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.














 DownLoad:
DownLoad: