-
Dear Editor,
On July 2, 2015, four pigs with severe respiratory distress and high fever (> 40.0 ℃) at a farm in Jeonpook province in the Southern region of South Korea were euthanized, and their lung tissues were collected and homogenized in PBS. The homogenized samples were inoculated into the wells of 6-well plates containing MadinDarby Canine Kidney (MDCK) cells in MEM with 1 μg/mL trypsin. One influenza A virus, A/Swine/Korea/ S2001/2015 (H3N2), was isolated from one sample. Moreover, a novel H3N2 influenza virus containing the matrix (M) gene from a 2009 pandemic influenza virus was detected in Korean pigs in 2013 (Pascua et al., 2013), an H1N2 influenza virus containing the internal genes from a 2009 pandemic influenza virus was found in Korean pigs in 2014 (Kim et al., 2014), and an H1N1 influenza virus containing all genes from the classical swine influenza viruses was isolated from Korean pigs in 2007 (Song et al., 2007).
On July 2, 2015, four pigs with severe respiratory distress and high fever ( > 40.0 ℃) at a farm in Jeonpook province in the Southern region of South Korea were euthanized, and their lung tissues were collected and homogenized in PBS. The homogenized samples were inoculated into the wells of 6-well plates containing Madin-Darby Canine Kidney (MDCK) cells in MEM with 1 mg/mL trypsin. The presence of influenza A virus in the inoculated wells was confirmed by reverse transcription polymerase chain reaction (RT-PCR) with influenza A-specific matrix (M) primers (available upon request). One influenza A virus, A/Swine/Korea/S2001/2015 (H3N2), was isolated from one sample. Four different lung samples of pigs were confirmed by PCR with influenza M gene-specific primers, and theviruses were then isolated from MDCK cells. Onevirus was grownin MDCK cells. Additional sequences of RNAs isolated from three lung samples that were not successfully isolated from MDCK cells were similar to those of the virus isolated from MDCK cells. Alleight genes of the isolate were sequenced and deposited the sequences in GenBank (accession numbers: KT427453-KT427460).
A BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi, http://platform.gisaid.org/epi3/frontend#4ead5c) was performed to identify sequences closest to those of previously isolated H3N2 influenza viruses (Table 1). The RNA polymerase basic subunit 2 (PB2), RNA polymerase basic subunit 1 (PB1), hemagglutinin (HA), and neuraminidase (NA) genes were closely related to those of the Korean swine H3N2 and H3N1 influenza viruses A/swine/Korea/CY03-19/2012(H3N2), A/swine/Korea/CY02-08/2A/2012(H3N1), A/swine/Korea/KSB/2012(H3N2), and A/swine/Korea/CY02-09/2012(H3N2), respectively, with over 98% identity. The genes encoding RNA polymerase acidic subunit (PA), nucleoprotein (NP), and M were closely related to those of the 2009 pandemic H1N1 influenza viruses A/Singapore/GP1146/2009(H1N1), A/Uganda/MUWRP-093/2009(H1N1), and (A/Hawaii/29/2009(H1N1), respectively, with over 98% identity. The nonstructural (NS) gene was closely related to that of the Korean swine H1N2 influenza virus A/swine/Korea/JL02/2005 (H1N2), with 97% identity.
Viral genes Closest related viruses Nucleotide identity (%) PB2 A/swine/Korea/CY03-19/2012(H3N2) 99 PB1 A/swine/Korea/CY02-08/2A/2012(H3N1) 99 PA A/Bangkok/SIMI508/2010(H1N1) A/Singapore/GP1146/2009(H1N1) Scotland/Edinburgh_20204/2009(H1N1) 98 HA A/swine/Korea/CY03-13/2012(H3N1) A/swine/Korea/KSB/2012(H3N2) 99 NP A/Athens/INS413/2010(H1N1) A/Uganda/MUWRP-093/2009(H1N1) A/Tennessee/F1052A/2010(H1N1) 98 NA A/swine/Korea/CY02-09/2012(H3N2) 98 M A/Tennessee/F1078/2010(H1N1))(A/Hawaii/29/2009(H1N1) A/Mexico City/INER16/2009(H1N1) 99 NS A/swine/Korea/JL02/2005(H1N2) 97 PB2:RNA polymerase basic subunit 2; PB1: RNA polymerase basic subunit 1; PA: RNA polymerase acidic subunit; HA: hemagglutinin; NP: nucleoprotein; NA: neuraminidase; M: matrix gene; NS: nonstructural gene. Table 1. Nucleotide homology of A/Swine/Korea/S2001/2015 (H3N2) with the closest related influenza viruses
Phylogenic analysis showed that five genes (PB2, PB1, HA, NA, and NS) of our isolate belonged to the swine lineage (Supplementary Figure S1A, S1B, S1D, S1F, and S1H), and the remaining genes (PA, NP, and M) were clustered with the human lineage (Supplementary Figure S1C, S1E, and S1G).
Next, amino acid sequences of the isolated swine H3N2 influenza viruswere analyzed. Positions 138 and 228 of HA protein (H3 numbering) contained an alanine (A) and a serine (S) residue, respectively, which were previously found to be related to enhanced binding to the human influenza receptor (α2-6-linked sialic acids) compared with that to the avian influenza receptor (α2-3-linked sialic acids)(Wang et al., 2010). The HA protein had a valine (V) at position 226 instead of a leucine (L), as is found in many human H3N2 influenza viruses. The NA protein had arginine (R), arginine (R), and histidine (H) residues at positions 152, 274, and 292, respectively, instead of lysine (K), lysine (K), and tyrosine (Y) residues, which are related to resistance to the anti-influenza drugs oseltamivir and zanamivir (Tamura et al., 2009). Alanine (A) was present in M1 at position 215, suggesting increased virulence in mice (Fan et al., 2009). NS1 harbored a serine (S) at position 42, which is related to enhanced pathogenesis in mice (Jiao et al., 2008). Asparagine (N) was identified at position 31 of M2; this residue has been shown to confer resistance to the anti-influenza drugs amantadine and rimantadine (Hay et al., 1985).
Three-week-old pigs (n = 6 per group), which did not harbor any antibodies against swine H1 and H3 subtypes of influenza viruses were intranasally (i.n.) infected with 1 mL of 106 TCID50/mL of A/Swine/Korea/S2001/2015 (H3N2) to determine the clinical signs of infection, such as such as the changes of in body temperature and weight, and to the evaluate pathological damage to the lungs. The infected pigs (n = 5) exhibitedsneezing and running noses; however, no pigs died as a result of infection. Pigs started to show increased body temperature and loss of body weight starting 2 days p ost infection (p.i.), but recovered around 8 days p.i. (Figure 1). When we measured body temperatures, the highest temperature (39.6 ℃) in the infected pigs was observed at 4 days p.i. (Figure 1A). Infected pigs lost as much as 15.2% of their body weight by 6 days p.i. (Figure 1B). Additionally, measurement of viral titers from nasal swabs collected in 1mL PBS from 1-10 days p.i. showed that peak viral titers (5.0 log10TCID50/mL) were observed at 4 days p.i. (Figure 1C). One infected pig and one uninfected pig were euthanized at 5 days p.i. for pathological staining (Figure 2). When the lung tissues were stained with hematoxylin and eosin (H & E) the infected lungs showed severe interstitial pneumonia with major infiltration of inflammatory cells in contrast to that of the uninfected pig. Lung tissues were also stained with anti-influenza A nucleoprotein (NP) antibodies and Fast Red to confirm the lung infection induced by our isolate. Many positive cells were detected in cells of the bronchi and bronchioles in infected lung tissues; in contrast, no positive staining was observed in uninfected lung tissues.
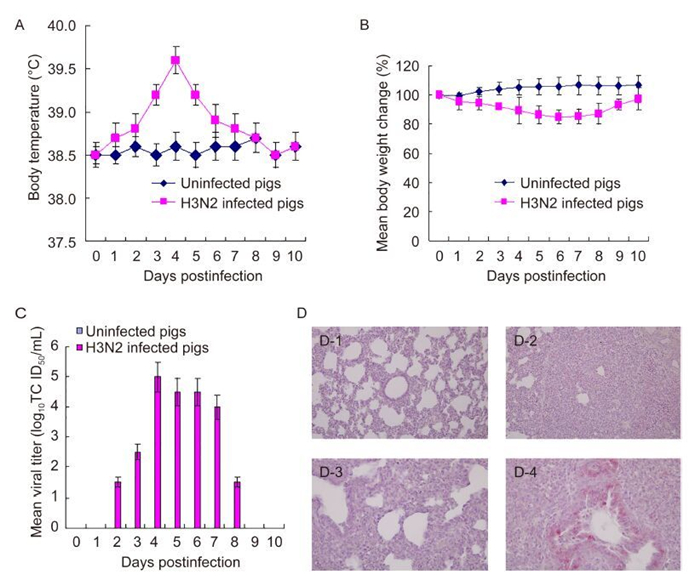
Figure 1. Clinical signs of pigs. Healthy 3-week-old pigs (n = 5 per group) were intranasally infected with 1 mL of 106TCID50/mL A/Swine/Korea/S2001/2015 (H3N2) and were checked for changes in body temperature and weight. Viral titers in nasal swabs were measured. Data are the means± standard deviations of five pigs. (A), changes in body temperature; (B), changes in body weight; (C), viral titers in nasal swabs. Lung tissue staining of infected pigs. Three-week-old healthy pigs (n = 1 per group) were intranasally infected with 1mL of 106 TCID50/mL A/Swine/Korea/S2001/2015 (H3N2). (D) Five days later, pigs were euthanized, and lung tissues were collected. The lung tissues were stained with hematoxylin and eosin (D-1 and D-2, 200 ×) or with rabbit anti-influenza A antibodies and Fast Red with hematoxylin counterstaining (D-3 and D-4, 400 ×). D-1 and D-3, lung tissues from uninfected pigs; D-2 and D-4, lung tissues from infected pigs.
To the best of our knowledge, this is the first report of the circulation of a novel reassortant swine H3N2 influenza virus, A/Swine/Korea/S2001/2015 (H3N2), containing genes from swine H3N2 or H3N1, 2009 pandemic H1N1, and Korean swine H1N2 influenza viruses in Korean pigs. The findings of this novel strain suggest that pigs could act as a host for reassortant influenza viruses. Previous studies have reported that swine H3N2 and H3N1 influenza viruses containing the M gene from the 2009 pandemic H1N1 influenza virus and 2009 pandemic H1N1 influenza virus circulated in Korean pigs in 2012 (Pascua et al., 2013). When we surveyed other pigs on the same farm from which the novel reassortant H3N2 influenza virus was isolated, revealed the presence of one genetically similar H3N2 influenza virus from 10 nasal swabs of pigs suffering from respiratory distress.
Notably, swine H3N2 influenza viruses containing the PA, NP, and M genes of the 2009 pandemic H1N1 influenza virus were found in 2013 in Vietnam (Baudon et al., 2015). The other internal genes (PB2, PB1, and NS) of swine H3N2 influenza viruses found in 2013 in Vietnam were derived from swine H3N2 influenza viruses (Baudon et al., 2015); however, the NS gene of our isolate was derived from a swine H1N2 influenza virus circulating in Korean pigs. In China, novel triple reassortant H1N2 influenza viruses containing PB2, PB1, PA, NP, M, and NS genes of 2009 pandemic H1N1 influenza viruses were detected in pigs in 2012 (Qiao et al., 2014), and novel triple-reassortant H1N1 influenza viruses containing PB1, PB1, PA, and NP of 2009 pandemic H1N1 influenza viruses were found in pigs from 2009 to 2013 (Sun et al., 2016). In Japan, H1N2 influenza viruses containing PB2, PB1, PA, M, and NS genes of 2009 pandemic H1N1 influenza viruses and H3N2 influenza viruses containing PB2, PB1, PA, NP, M, and NS genes of 2009 pandemic H1N1 influenza viruses were isolated from pigs in 2013 (Kanehira et al., 2014). In humans, 2009 pandemic H1N1, H3N2, and human B have been circulating in China and Korea since 2009 (Guan et al., 2015; Kim et al., 2011). Our isolate, A/Swine/Korea/S2001/2015 (H3N2), could replicate in pigs and cause severe pneumonia in infected pigs. Similarly, a previous study (Rajão et al., 2015) showed that a similar human-like H3N2 influenza (A/Swine/Missouri/A01476459/2012) with internal genes of the 2009 pandemic H1N1 influenza virus replicated in pigs and caused severe pneumonia in infected pigs.
One possible explanation for the novel H3N2 influenza viruses occurring in pigs in Asian countries is that farm workers infected with the 2009 pandemic H1N1 influenza virus came in contact with pigs infected with swine H3N2 influenza viruses, resulting in the generation of novel H3N2 influenza viruses containing genes from the 2009 pandemic H1N1 viruses. Pigs are considered a "mixing vessel", permitting the generation of new reassorted influenza A viruses since they contain human and avian influenza receptors (Ito et al., 1998).
In summary, our results suggest that close monitoring of this novel H3N2 influenza virus in pigs and humans is required to prevent crossover infection with this virus in humans.
HTML
-
This study was in part funded by a 2015 research fund from Chungnam National University. The authors declare that they have no conflicts of interest. The study was approved by the Institutional Animal Care and Use Committee of Chungnam National University.
Supplementary figure is available on the websites of Virologica Sinica: www.virosin.org; link.springer.com/journal/12250
-
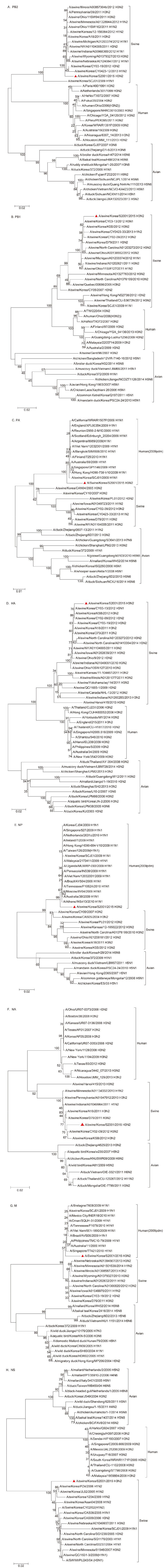
Figure S1. Phylogenetic analysis of PB2, PB1, PA, HA, NP, NA, M, and NS genes of A/Swine/Korea/S2001/2015 (H3N2). The phylogenetic tree was constructed using the neighbor-joining method in Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 (Tamura et al, 2007) with 1000 bootstrap replicates. The scale bar shows nucleotide substitutions per site. (A), PB2; (B), PB1; (C), PA; (D), HA; (E), NP; (F), NA; (G), M; (H), NS.
















 DownLoad:
DownLoad: