HTML
-
Actin exists in two forms: monomeric globular actin (G-actin) and filamentous actin (F-actin). In most cases, G-actin polymerization to F-actin requires actin-related protein complex (Arp2/3), which is composed of seven subunits: Arp3, Arp2, p40/ARPC1 (P40), p34/ARPC2 (P34), p21/ARPC3 (P21), p20/ARPC4 (P20), and p16/ARPC5 (P16) (Welch and Mullins, 2002; Bovellan et al., 2014). The activation of Arp2/3 is heavily dependent on nucleation promoting factors (NPFs), which usually harbor a conserved VCA domain that interacts with and activates Arp2/3 (Machesky and Insall, 1999; Rohatgi et al., 1999; Winter et al., 1999).
The actin cytoskeleton is actively involved in a variety of cellular processes ranging from cell mobility and vesicle trafficking to transcription regulation and chromatin remodeling (Svitkina et al., 1997; Svitkina and Borisy, 1999a; Svitkina and Borisy, 1999b; Olave et al., 2002; Percipalle et al., 2003). Viral pathogens also harness the host actin cytoskeleton to facilitate their replication. Alphabaculoviruses are well-documented viral pathogens that induce host cell actin polymerization in a separate and sequential manner (Charlton and Volkman, 1991; Charlton and Volkman, 1993). Upon viral entry into insect cells, F-actin cables are immediately formed to assist in viral migration from the cytoplasm into the nucleus (Charlton and Volkman, 1993; Ohkawa et al., 2010). After the virus enters the nucleus, nuclear actin polymerization occurs and F-actin participates in viral nucleocapsid assembly (Ohkawa and Volkman, 1999; Goley et al., 2006).
Virus-induced nuclear actin polymerization requires key elements of the cytoplasmic actin polymerization machinery, including G-actin, NPFs, and Arp2/3, to relocate to the nucleus. Autographa californica multiple nucleopolyhedrovirus (AcMNPV), one of the best-characterized alphabaculoviruses, utilizes six early viral genes to translocate cytoplasmic G-actin into the nucleus at the early infection phase (Ohkawa et al., 2002; Gandhi et al., 2012). P78/83, an AcMNPV-encoded NPF, is relocated to the nucleus by the viral nucleocapsid protein BV/ODV-C42 (Wang et al., 2008; Li et al., 2010). We have identified that the viral late-gene product Ac34 mediates the nuclear entry of Arp2/3 P40 (Mu et al., 2016).
Ac34 is highly conserved among all alphabaculoviruses sequenced to date. Knockout of ac34 from the AcMNPV genome results in attenuated viral replication in Spodoptera frugiperda Sf9 cells (Cai et al., 2012), or complete replication deficiency in Sf21 cells (Salem et al., 2013). The significance of Ac34 in AcMNPV replication may be attributed (but not limited) to its role in the nuclear relocation of P40, one of the subunits of Arp2/3. However, it remains unknown whether the other subunits of Arp2/3 are also relocated to the nucleus by Ac34, and whether Ac34 plays a universal role to relocate mammalian Arp2/3.
In this study, we continued to explore the role of Ac34 in the nuclear relocation of Arp2/3 subunits and attempted to reveal the molecular mechanism underlying the different capabilities of Ac34 in nuclear relocating Arp2/3 from different species.
-
Sf9 cells from Spodoptera frugiperda were cultured in Grace's medium (Invitrogen, Carlsbad, USA) with 5% fetal bovine serum (FBS; Gibco, Australia Origin) and 0.1% antibiotic-antimycotic (Invitrogen) at 27 ℃. Sf9 cells were transfected with the indicated plasmid (see below), using Cellfectin Ⅱ reagent (Invitrogen) by following standard procedures.
293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% FBS (Gibco) and 1% penicillin and streptomycin. 293T cells were transfected with the indicated plasmid (see below), using Lipofectamine 2000 (Invitrogen) by following standard procedures.
-
Total RNA was extracted from Sf9 cells or 293T cells by using TRIzol reagent (Invitrogen), and was converted to cDNA by reverse transcription using oligo (dT) and M-MLV reverse transcriptase (Promega, Fitchburg, WI, USA), according to the manufacturer's protocols. Gene-specific primers for the Arp2/3 subunits of S. frugiperda or Homo sapiens were designed according to the SPODOBASE (http://bioweb.ensam.inra.fr/spodobase/) or GenBank (http://www.ncbi.nlm.nih.gov/genbank/) database, and were used to amplify the open reading frame (ORF) of individual Arp2/3 subunits by polymerase chain reaction. The ORF of individual Arp2/3 subunits was inserted into pIZ-V5/HA (for expression in Sf9 cells) or pXJ40-HA (for expression in 293T cells). Other plasmid constructs encoding Ac34 fusion proteins were constructed based on standard protocols. The primers used in this study are listed in Table 1.
Name Sequence (5′ to 3′) Restriction sites Sf-p40-S CGGGGTACCAAGATGTCTCAAACTCTTACATTCGG Kpn Ⅰ Sf-P40-A CGCGGATCCAACAATCTTCAGTCCCTCAATG BamH Ⅰ Sf-p20-S CCCAAGCTTATGTCGGCTACATTAAAACCT Hind Ⅲ Sf-p20-A CGCGGATCCGAATCTCTTCAAAAACTCTT BamH Ⅰ Sf-p16-S CCCAAGCTTATGGCGAAAAACACGTCGAGT Hind Ⅲ Sf-p16-A CCGAGCTCCACTTTCATTCGGTTCA Sac I Sf-p34-S CCCAAGCTTATGATCTTACTCGAGA Hind Ⅲ Sf-p34-A CGCGGATCCGTCTCGCCTCACAAATGTTCT BamH Ⅰ Sf-Arp2-A CGCGGATCCACTGGCGCGAGGACCCA BamH Ⅰ Sf-Arp2-S CCCAAGCTTATGGATGAGAAAGGAAGA Hind Ⅲ mCherry-S CCGGAATTCATGGTGAGCAAGGGCGAGGA EcoR Ⅰ Ac34-S CGCGGATCCATGACAACGGTTGCTGTG BamH Ⅰ Ac34-A CCGGAATTCTTACTCAAAGTCCATCAATTCGTAC EcoR Ⅰ Ac34-myc-S CGCGGATCCATGACAACGGTTGCTGTG BamH Ⅰ Ac34-myc-A CCGGAATTCTTACTCAAAGTCCATCAATTCGTAC EcoR Ⅰ Hs-p40-S CCCAAGCTTATGGCCTACCACAGCTTCCT Hind Ⅲ Hs-p40-A CGGGGTACCTCATTTGATCTTGAGGTCCTTCAA BamH Ⅰ Hs-p34-S CCCAAGCTTATGATCCTGCTGGAGGTGAAC Hind Ⅲ Hs-p34-A CGGGGTACCTTAGCGGGATGAAAACGTCTTC BamH Ⅰ Hs-p21-S CCCAAGCTTATGCCGGCTTACCACTCTTC Hind Ⅲ Hs-p21-A CGGGGTACCTCACTGTCCAGGTCCTGAAAG BamH Ⅰ Hs-p20-S CCCAAGCTTATGACTGCCACTCTCCGCC Hind Ⅲ Hs-p20-A CGGGGTACCTTAAAAATTCTTAAGGAACTCTTCAGC / Hs-p16-S CCCAAGCTTATGTCGAAGAACACAGTGTCGT Hind Ⅲ Hs-p16-A CGGGGTACCCTACACAGTTTTTCTTGCAGTCAA BamH Ⅰ Hs-Arp3-S CCCAAGCTTATGGCGGGACGGCTGCC Hind Ⅲ Hs-Arp3-A CGGGGTACCTTACGACATGACTCCAAACACTG BamH Ⅰ Hs-Arp2-S CCGCTCGAGATGGACAGCCAGGGCAGG / Hs-Arp2-A CGGGGTACCTTATCGAACAGTCACACCAAGTTT / Table 1. Sequences of primers used in this study
-
An immunoprecipitation (IP) assay was performed as described previously (Wang et al., 2015). Briefly, the cells were rinsed with ice-cold PBS and lysed with RIPAbuffer (50 mmol/L Tris, pH = 7.5, 1 mmol/L EGTA, 1 mmol/L EDTA, 1% Triton X-100, 150 mmol/L NaCl, 2mmol/L DTT, 100 µmmol/L PMSF, 1 µg/ml Proteinase Inhibitors).The cell lysates were centrifuged at 20, 817 × g at 4 ℃ for 10 min, and the supernatants (whole-cell lysate, WCL) were collected. The protein concentration of the WCL was determined by Bradfordassays, and 1500 µg total protein was mixed with 2 µg anti-HA or anti-FLAG (Sigma, Shanghai, China) and protein-G agarose (Millipore, Billerica, Massachusetts, USA), andincubated at 4℃ overnight. The immunoprecipitatedsamples were centrifugedand washedthree times, and subjected to a western blot assay using anti-HA, anti-FLAG, or anti-mCherry (1:1000 dilution, Invitrogen).
-
The immunofluorescence staining was performed as described previously (Wang et al., 2008). Briefly, cells were fixed with 3.7% paraformaldehyde in PBS for 30 min, permeabilized with 0.5% TritonX-100, and blocked in 1% normal goat serum (Boster Biological Technology, wuhan, China) in PBS for 30 min on ice. The cells were incubated with the following primary antibodies: anti-myc (1:1000 dilution, Santa Cruz Biotechnology, Texas 75220, USA) or anti-HA (1:1000 dilution, Sigma). The secondary antibodies were Alexa Fluor 488-conjugated anti-mouse or anti-rabbit (1:800 dilution, Invitrogen). The nuclear DNA was stained with Hoechst 33258 (Beyotime, Shanghai, China).
Fluorescence intensities of individual Arp2/3 subunits in the whole cell or the nuclear region were measured using Volocity software (Perkin Elmer). At least 50 positively transfected cells from three independent experiments were subjected to a densitometry assay. The Student's t-test was performed to compare the relative nuclear fluorescence intensities in the absence or presence of Ac34.
Cell culture and transfection
Cloning of Arp2/3 subunits and plasmid construction
Immunoprecipitation
Immunofluorescence staining and densitometry assay
-
We have identified that the AcMNPV late-gene product Ac34 induces the nuclear relocation of P40 of Arp2/3 during AcMNPV infection (Mu et al, 2016). In order to test the influence of Ac34 on the other Arp2/3 subunits, Arp2, Arp3, p34, p20, and p16 were cloned from Sf9 cells and their sequence information was submitted to GenBank (Accession numbers: AFD50557 (P40) (Han et al., 2012); ADB27912 (P21) (Li et al., 2010); Arp3: in submission; AHV90274 (Arp2); AHV90277 (P16); AHV90276 (P20); AHV90275 (P34)).
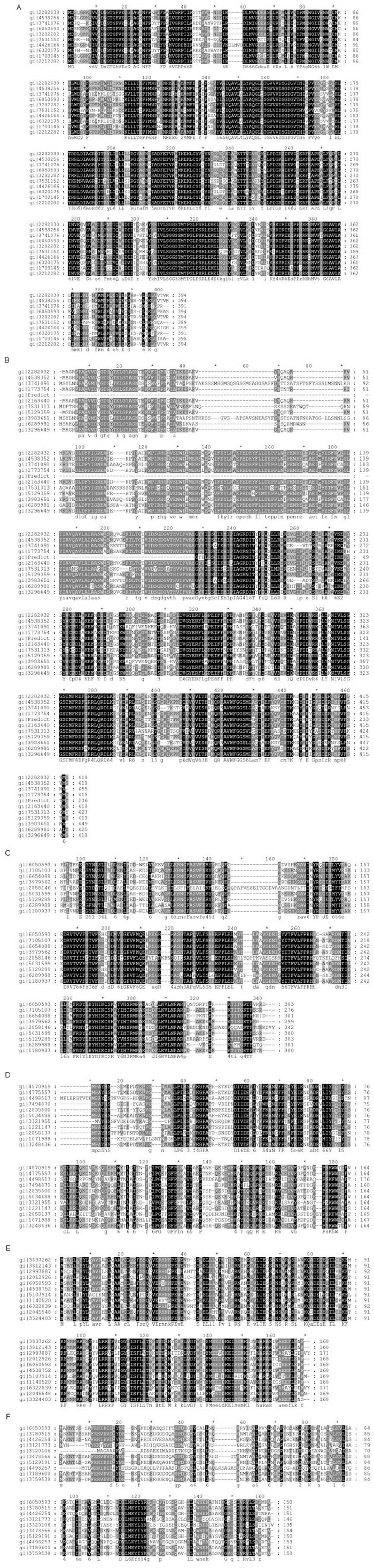
Amino acid sequences of Arp2/3 subunits from different species, including S. frugiperda, H. sapiens, Ashbya gossypii, Arabidopsis thaliana, Bombyx mori, Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, Gallus gallus, Mus musculus, Saccharomyces cerevisiae, and Schizosaccharomyces pombe, were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) (Supplementary Figure S1). In particular, P16 showed the lowest similarity, whereas Arp2 exhibited the highest similarity among all the species included in the alignment.
-
Previously, we identified that the viral protein Ac34 induced P40 nuclear relocation during AcMNPV infection of Sf9 cells (Mu et al., 2016). To explore further the role of Ac34 in nuclear relocation of Arp2/3, we compared the subcellular distribution of other Arp2/3 subunits in the absence or presence of Ac34 by a quantitative fluorescence assay. In the absence of Ac34, HA-tagged P40, P20, P34, Arp2, and Arp3, and red fluorescent protein-tagged P21 showed a predominantly cytoplasmic distribution pattern (Figure 1A-F). When co-expressed with mCherry-tagged Ac34 (mC-Ac34), epitope-tagged P40, P21, P20, and P34 showed significant nuclear relocation (Figure 1A-D), whereas Arp2 and Arp3 displayed no spatial changes (Figure 1E, F), as indicated by a densitometry assay. In contrast to the other Arp2/3 subunits, a small fraction of P16 was also localized in the nucleus in the absence of Ac34(Figure 1G), which may be attributed to the free nucleocytoplasmic shuttling of low-molecular-weight proteins, and implied that the nuclear relocation mechanism of Arp2, Arp3, and P16 might vary from that of other Arp2/3 subunits that are heavily dependent on Ac34. As a control, mC-Ac34 and mCherry were expressed in Sf9 cells, and showed a similar subcellular distribution pattern to when they were co-expressed with individual Arp2/3 subunits (Figure 1H).
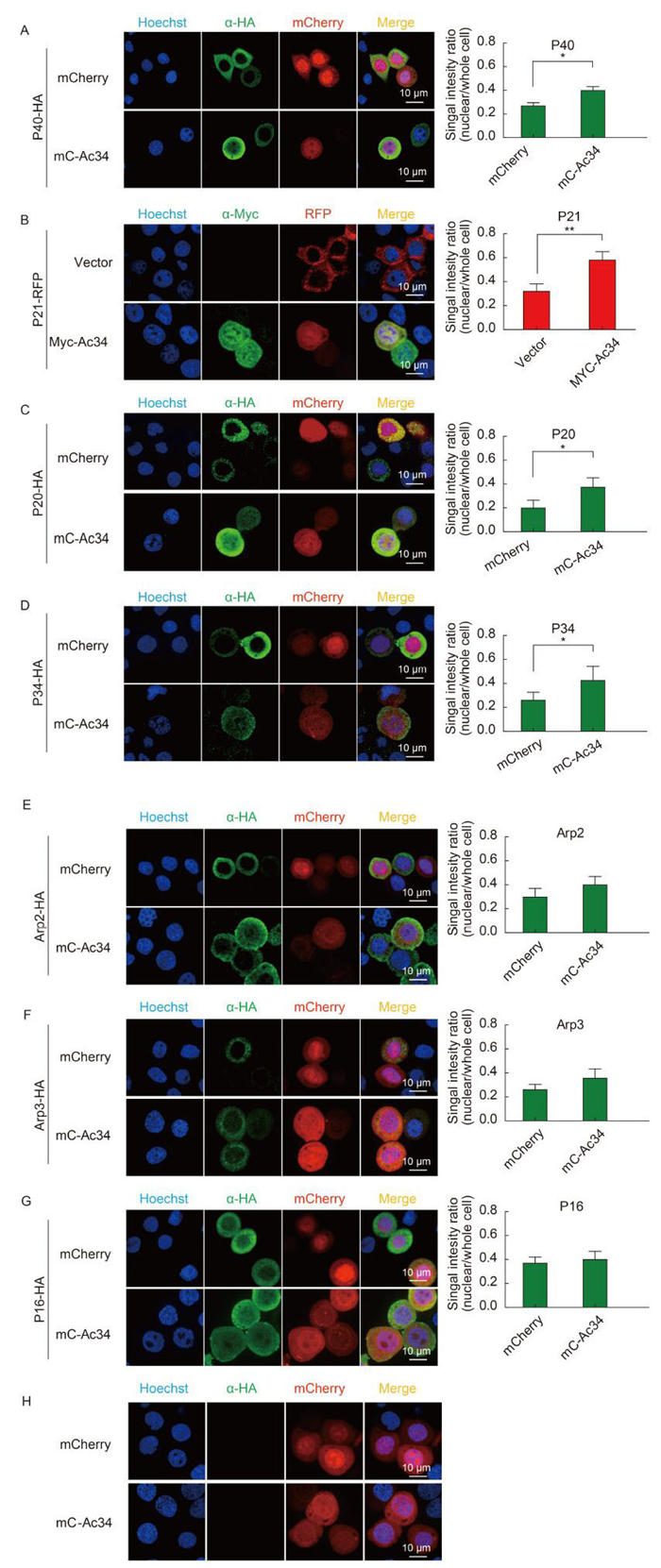
Figure 1. Ac34 has different impacts on the subcellular distribution pattern of S. frugiperda Arp2/3 subunits. (A-G) S. frugiperda Arp2/3 subunits were co-expressed with mCherry or Ac34 fusion protein in Sf9 cells. At 48 hours post-transfection (hpt), cells were analyzed by immunofluorescence microscopy. Fluorescence intensities of individual Arp2/3 subunits in the whole cell or the nuclear region were measured with Volocity software (Perkin Elmer). At least 50 positively transfected cells from three independent experiments were subjected to a densitometry assay. The Student's t-test was performed to compare the relative nuclear fluorescence intensities in the absence or presence of Ac34. *, P < 0.05; **, P < 0.01. (H) Subcellular distribution of mCherry and mC-Ac34. mCherry and mC-Ac34 were expressed in Sf9 cells, and immunofluorescence microscopy was performed at 48 hpt. The images were captured using a Perkin Elmer UltraVIEW VoX microscope.
-
Because Ac34 plays an active role in the nuclear relocation of S. frugiperda Arp2/3 subunits, we continued to explore whether Ac34 plays a universal role in the nuclear relocation of Arp2/3 subunits from different species. HA-tagged Arp2/3 subunits of H. sapiens were co-expressed with mCherry or mC-Ac34, respectively. Immunofluorescence microscopy and a densitometry assay demonstrated that all subunits of Arp2/3 were mainly distributed in the cytoplasm, regardless of the presence of Ac34 (Figure 2). The localization patterns of Arp2, Arp3, and P34 were in accordance with previous reports (Welch et al., 1997; Yoo et al., 2007).
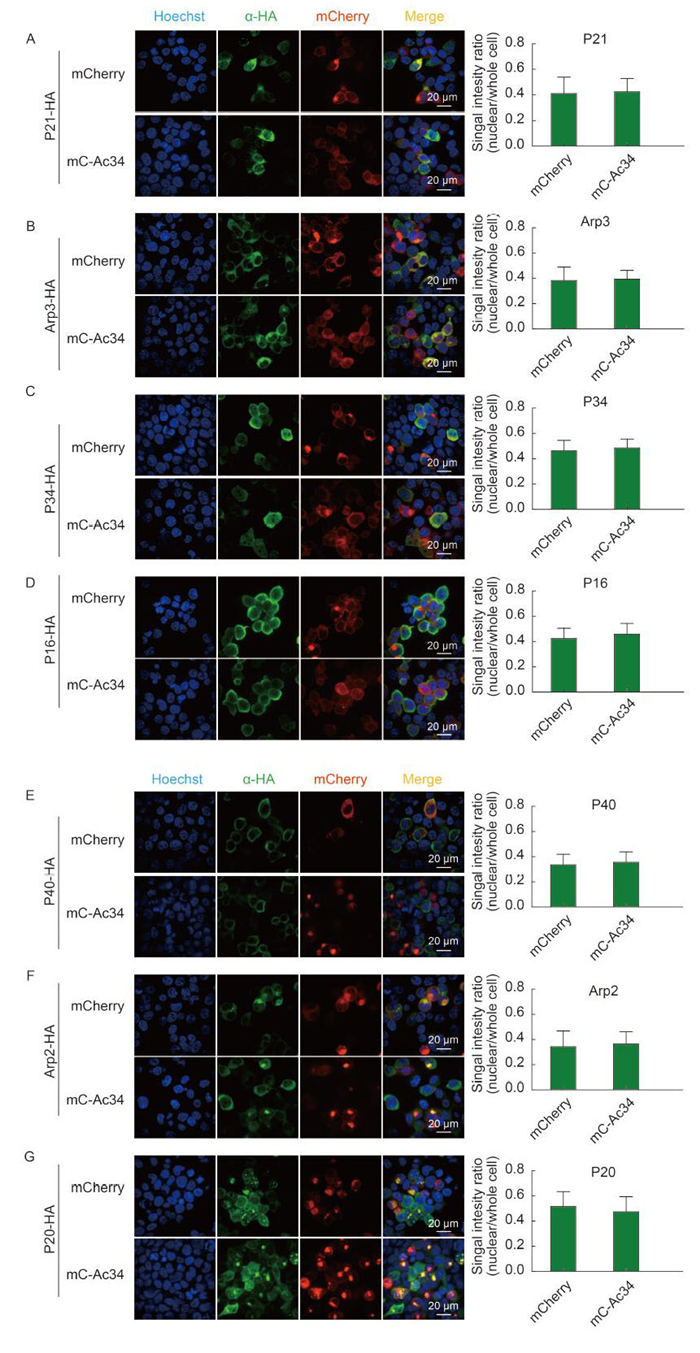
Figure 2. Ac34 has no impact on the subcellular distribution of H. sapiens Arp2/3 subunits. (A-G) H. sapiens Arp2/3 subunits were co-expressed with mCherry or mC-Ac34 in 293T cells. At 48 hours post-transfection (hpt), cells were analyzed by immunofluorescence microscopy. Fluorescence intensities of individual Arp2/3 subunits in the whole cell or the nuclear region were measured with Volocity software (Perkin Elmer). At least 50 positively transfected cells from three independent experiments were subjected to a densitometry assay. The Student's t-test was performed to compare the relative nuclear fluorescence intensities in the absence or presence of Ac34.
-
Previously, we have demonstrated that Ac34 interacts with P40 of S. frugiperda, and that this interaction is required for P40 nuclear relocation during AcMNPV infection of Sf9 cells (Mu et al., 2016). In this study, we demonstrated that Ac34 exhibits different capabilities in the nuclear relocation of different Arp2/3 subunits. One of the possible explanations contributing to this difference is that Ac34 possesses different binding capabilities to different Arp2/3 subunits, which leads to different levels of nuclear relocation.
To test the interaction between Ac34 and different Arp2/3 subunits, mC-Ac34 was co-expressed with the different Arp2/3 subunits, and co-IP was performed to analyze the interactions. As shown in Figure 3, the immunoprecipitated P40 generated a strong band in the western blot, indicating a strong interaction between Ac34 and P40; in addition, the immunoprecipitated P21, P34, and P20 generated weak bands in comparison with P40, implying relatively weak interactions with Ac34. Precipitation of Arp3 and P16 failed to generate visible bands, indicating that Ac34 does not interact with these two Arp2/3 subunits. Precipitation of Arp2 also generated a weak band at approximately 35 kDa, failing to match the molecular weight (MW) of Arp2 (45 kDa). These phenotypes are in accordance with the immunofluorescence assay results showing that Ac34 failed to relocate Arp2, Arp3, and P16 to the nucleus, suggesting that an interaction with Ac34 is essential for Arp2/3 subunits to relocate to the nucleus. Note that all the subunits except for P40 showed low protein abundance when expressed in Sf9 cells, which could also be responsible for the weak bands observed in the co-IP assay.
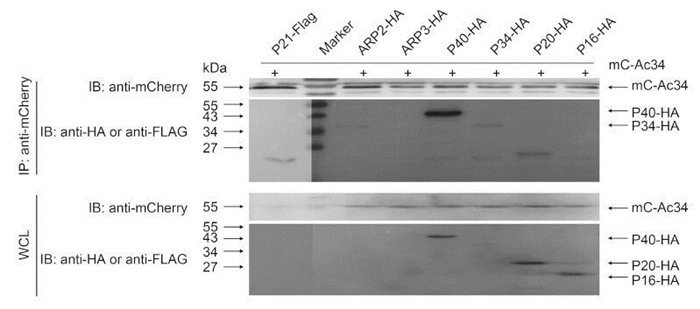
Figure 3. Interaction between Ac34 and Arp2/3 subunits of S. frugiperda. mC-Ac34 and FLAG-or HA-tagged Arp2/3 subunits of S. frugiperda were co-expressed in Sf9 cells. A co-immunoprecipitation assay using anti-mCherry was performed at 48 hours post-transfection (hpt) and precipitated proteins were probed with the indicated antibodies.
Not surprisingly, all of the Arp2/3 subunits of H. sapiens failed to interact with Ac34 (Figure 4), providing further evidence that an interaction with Ac34 is essential for the nuclear relocation of Arp2/3 subunits, irrespective of the cell's origin (insect or mammal).
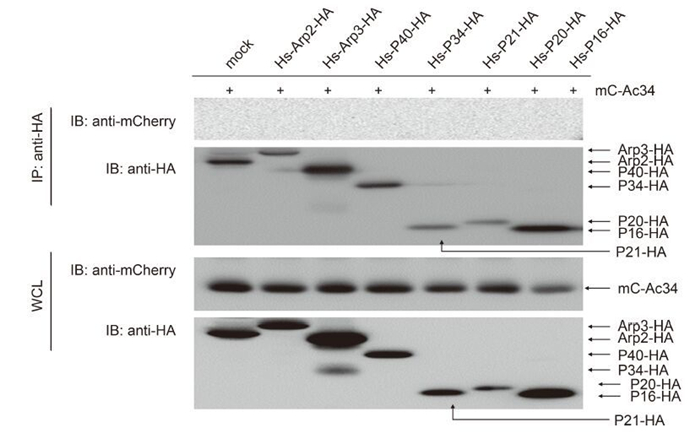
Figure 4. Interaction between the Ac34 and Arp2/3 subunits of H. sapiens. mC-Ac34 and HA-tagged Arp2/3 subunits of H. sapiens were co-expressed in 293T cells. A co-immunoprecipitation assay using anti-HA was performed at 48 hours post-transfection (hpt) and the precipitated proteins were probed with the indicated antibodies.
Sequence of Arp2/3 subunits of S. frugiperda
Ac34 showed different capabilities with respect to nuclear relocation of different Arp2/3 subunits
Ac34 had no influence on the subcellular distribution of H. sapiens Arp2/3
Interaction of the Ac34 and Arp2/3 subunits
-
Arp2/3 plays an important role in a variety of biological processes that occur in either the cytoplasm or the nucleus. However, little is known about the Arp2/3 nuclear relocation mechanism. Alphabaculovirus infection induces nuclear actin polymerization in host cells, which requires cytoplasmic Arp2/3 to relocate to the nucleus. This unique feature has enabled the alphabaculovirus infection system to become a powerful research tool for investigating the mechanism of Arp2/3 nuclear relocation.
Arp2/3 is composed of seven subunits, and the MW is over 220 kDa. The results of our study demonstrated that not all subunits were recruited to the nucleus by Ac34. This phenotype suggested that each subunit employs a different mechanism to relocate to the nucleus. It is plausible to hypothesize that Arp2/3, as a high-MW-protein complex, may translocate to the nucleus as an individual subunit, to reduce the total MW and be able to pass through the nuclear pore complex more easily. In the nucleus, the individual Arp2/3 subunit would re-assemble into a functional protein complex to exert its activity in promoting actin polymerization.
In this study, we demonstrated that a protein-protein interaction is essential for Ac34 to relocate Arp2/3 subunits to the nucleus. One of the possible scenarios is that Ac34 contains a nuclear localization signal (NLS) and the Arp2/3 subunits bind to Ac34 and co-transport to the nucleus through the nuclear pore complex. Therefore, elucidating the detailed mechanism of Ac34 nuclear translocation would provide the key to understanding Arp2/3 nuclear relocation during AcMNPV infection. Our future research will focus on identifying the NLS of Ac34 and its role in the nuclear relocation of Arp2/3 subunits.
-
We thank Dr. Jan van Lent of Wageningen University and Dr. Ding Gao of core facilities in Wuhan Institute of Virology, CAS, for their technical assistance with fluorescence microscopy. This work was supported by grants from National Natural Science Foundation of China (31030027, 31470261, 31321001, and 31270191). The Royal Dutch Academy of Science and Arts (08-PSA-BD-01) is acknowledged for financing part of this research.
-
The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
-
MJF, WY, and CXW designed the experiments. MJF, ZYL, HX, and ZY carried out the experiments. MJF and HY analyzed the data. MJF, CXW and WY wrote the paper. All authors read and approved the final manuscript.
Supplementary Figure S1 is available on the websites of Virologica Sinica: www.virosin.org; link.springer.com/journal/12250
















 DownLoad:
DownLoad: