-
Porcine epidemic diarrhea virus (PEDV), a member of the genus Alphacornoavirus in the family Coronaviridae, causes porcine epidemic diarrhea (PED) and exhibits a tropism for epithelial cells of the small intestine (Song et al., 2012). Before 2013, PED was restricted to Asia (Song et al., 2012). However, after spring 2013, outbreaks of PEDV caused by variant PEDV strains occurred in North America, and researchers revealed that these strains might stem from Chinese variants (Huang et al., 2013).
The PEDV genome is a single-stranded positive-sense RNA genome that encodes four structural proteins: the envelope (E) glycoprotein, integral membrane (M) glycoprotein, nucleocapsid (N) protein, and spike (S) glycoprotein. The S protein plays an important role in receptor binding, membrane fusion, and induction of neutralizing antibodies, which makes it a suitable candidate for vaccine development.
With the help of furin-like proteases, the S protein is cleaved into the S1 (residues 1–789) and S2 (residues 790–1383) subunits (Sun et al., 2008). These two domains form the globular and stem region, respectively (Spaan et al., 1988; Duarte et al., 1994). Previous research has shown that the main epitopes are linear epitope domains S1D5 (residues 744–759) and S1D6 (residues 756–771), as well as a conformational region (residues 499–638), the collagenase-digested fragment (COE), located in the S1 domain. These epitopes are responsible for inducing virus neutralizing antibodies (Chang et al., 2002; Sun et al., 2008), making the S1 domain a potential target for vaccine design. Additionally, the variation in the S gene sequence is considered to contribute to antigenic diversity, so the S protein is also used in analyses of PEDV genotypes (Park et al., 2007).
The PEDV vaccine is currently the main measure of PEDV prevention and control. However, despite the fact that an inactivated vaccine has been widely used throughout China, the remarkable increase in the number of PEDV outbreaks since 2010 have caused significant economic losses and the effectiveness of the vaccine has therefore been questioned (Chen et al., 2012; Fan et al., 2012; Luo et al., 2012; Wei et al., 2012). Moreover, phylogenetic analysis of the S gene has revealed that the current field strains of PEDV in China represent a distinct branch compared to earlier field strains and the vaccine strains (Tian et al., 2013; Chen et al., 2013). In addition, neonatal piglets on many farms where the traditional inactivated vaccines were used experienced high mortality rates due to the pandemic PEDV strains. It is difficult to determine the effectiveness of the vaccines because of the lack of antibody surveillance in sows.
Although the detection of serum antibodies against a gastroenteric pathogen is only a sign of contact with the infectious microorganism, it has been suggested that the serum neutralizing (SN) titer could be used to determine herd infection status (Song et al., 2012). Using an enzyme-linked immunosorbent assay (ELISA) with PEDV particles as the coating antigen has also been proved to be reliable for detecting swine antibodies against PEDV (Hofmann et al., 1990; Oh et al., 2005). Whether these methods could be used to monitor the current PEDV epidemic requires further investigation.
In this study, we compared correlation coefficients between ELISA results (involving seven overlapping recombinant S fragments as well as the newly isolated PEDV strain, GDS01) and SN titers. Furthermore, ROC analyses were used to assess the reliability of the ELISAs.
-
The Vero cell line (ATCC CCL-81TM) was cultured at 27 °C in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and antibiotics (penicillin 100 U/mL and streptomycin 100 μg/mL) (Solarbio, Beijing, China). PEDV strain GDS01 (Genbank ID: KM089829.1), isolated in 2012 in Guangdong province, China, was propagated in Vero cells with 2.5 μg/mL trypsin and 10% tryptose phosphate broth solution (Sigma). A previous phylogenetic analysis of the S gene of GDS01 revealed that GDS01 clusters with other newly isolated strains ( Hao et al., 2014).
-
We used 15 PEDV-positive and 20 PEDV-negative serum samples to determine the optical density values at 450 nm (OD450) cutoff value in a checkerboard titration analysis of the ELISAs. The PEDV-positive serum samples were collected from PEDV-negative piglets that were experimentally infected with inactive GDS01. The serum samples were collected at day 7 after the booster immunization.
-
During 2012–2014, 156 sow serum samples were collected from four major Chinese swine-rearing provinces (Guangdong, Jiangsu, Guangxi, and Hunan), with known collection locations and gestation stages. The other background details of the samples were unknown. The 156 samples were used to assess the correlations between the SN titers and ELISA results (using different PEDV-specific antigens).
Briefly, 96-well plates (Griener, Frickenhausen, Germany) were coated with the relevant antigens at 4 °C overnight, and then blocked with 5% milk for 1 h. After washing three times with phosphate-buffered saline (PBS), 100 μL serum sample was added and the sample was incubated at 37 °C for 1 h. Subsequently, the plates were washed with PBS and incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Thermofisher, Waltham, Massachusetts, USA) at 37 °C for 1 h. The absorbance was measured at 450 nm.
-
To determine the antigenic regions to be used in the ELISAs, sequences of the S proteins of various PEDV strains were analyzed using DNAStar (version 7, Lasergene Corporation, Madison, WI, USA). The conformational region (COE in SP6) and linear epitopes (SS2 and SS6 in SP7) were also analyzed using DNAStar. Previous research identified a neutralizing epitope in the N-terminus of the S2 region in infectious bronchitis virus (IBV) (Koch et al., 1990). Thus, SP4, which is located at the N-terminus of S2, was selected according to the hydropathy and antigenicity plot predicated by the DNAStar software. To evaluate the entire S1 sequence, three other regions, SP1, SP3, and SP5, were also selected (Figure 1).
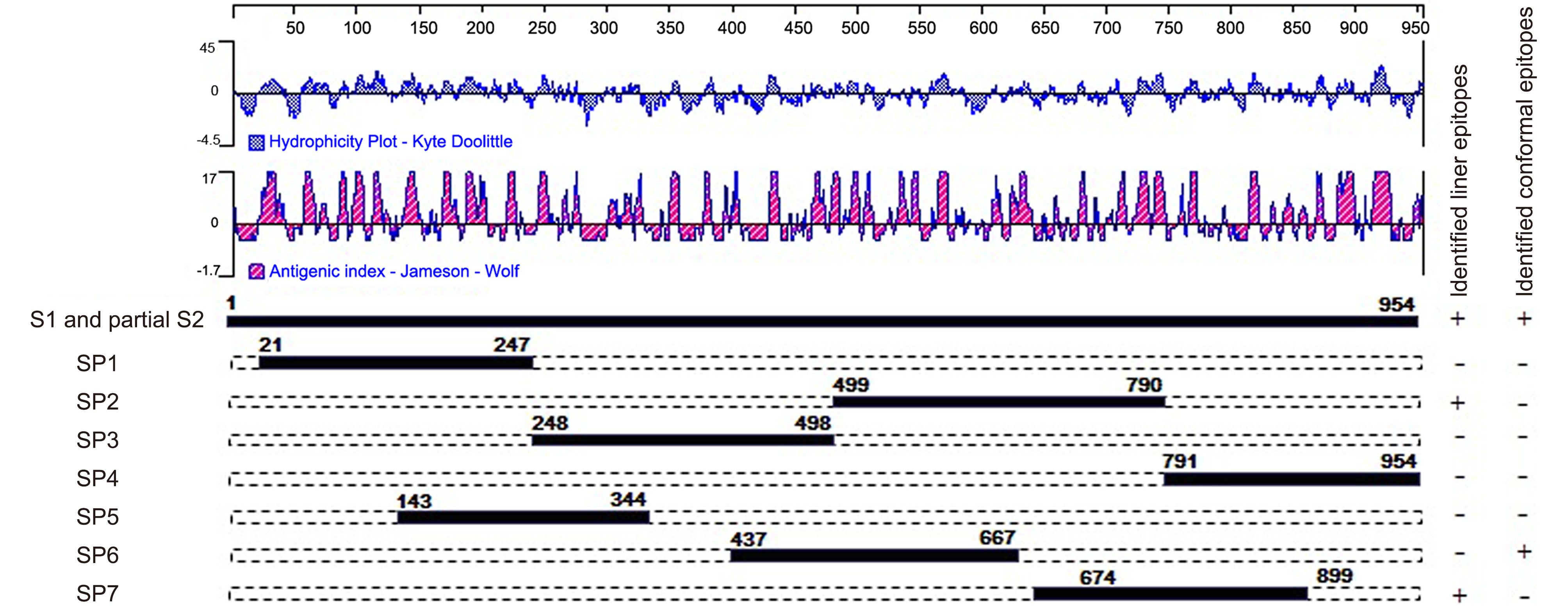
Figure 1. Schematic illustration of the location and length of the seven antigenic S fragments. The hydrophilicity plot and antigenic index were predicted by DNAStar software using the Kyte-Doolittle and Jameson-Wolf methods, respectively. The plus symbols indicate that the fragments contain identified neutralizing regions and the minus symbols indicate that the fragments do not contain identified neutralizing regions.
To construct the expression plasmids, seven overlapping cDNA clones containing S1 and partial S2 sequences from the GDS01 strain were amplified using different primer sets (Supplementary Table S1), with Kpn I and Xho I restriction enzyme sites. The amplified fragments were digested and then cloned into a pET32a plasmid (Novagen, Darmstadt, Germany). The resulting expression plasmids were labeled PEDV-S-SP1, PEDV-S-SP2, PEDV-S-SP3, PEDV-S-SP4, PEDV-S-SP5, PEDV-S-SP6, and PEDV-S-SP7.
-
The expression plasmids were transformed into Escherichia coli BL21 (DE3) competent cells. The protein expression and purification procedures were performed as described previously (Sun et al., 2008). Briefly, protein expression was induced by adding 1.0 mmol/L isopropyl β-D-1-thiogalactopyranoside (IPTG) and culturing at 16 °C for 24 h. After centrifuging, the bacterial cell pellets were dissolved in a 1/10 volume of PBS (0.01 mol/L, pH 7.2) and lysed by sonication. The inclusion bodies containing the majority of the recombinant S proteins were collected by centrifugation, and then suspended in binding buffer with a denaturing agent (20 mmol/L Na 3PO 4, 500 mmol/L NaCl, 30 mmol/L imidazole, and 8 mol/L urea).
All the recombinant proteins were expressed with a 6× polyhistidine tag (His-tag) at the C-terminus, and purified using a Ni-based resin column (GE Healthcare, Chicago, Illinois, USA) according to the manufacturer’s instructions. The purified recombinant proteins were renatured at 4 °C by step-wise dialysis against PBS (0.01 mol/L, pH 7.2) containing gradually decreasing concentrations of urea to remove the urea and imidazole. Both unpurified and purified recombinant S peptides were analyzed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) stained with Coomassie blue. The purified proteins were identified by western blotting using a monoclonal antibody against the His-tag (PTG, Manhattan, New York, USA). The protein concentration was determined by using the Bradford method (Bio-Rad, Hercules, California, USA).
-
PEDV virus propagation and purification were conducted as previously described (Hofmann et al., 1990). Briefly, Vero cells were washed twice with the PBS to remove the growth medium (DMEM supplemented with 10% heat-inactivated fetal calf serum and antibiotics). After cultivation for 60 min at 37 °C, the cells were washed with PBS. The washed cells were then infected with the virus by adding the “infection medium” (DMEM and 0.3% tryptose phosphate broth with 10 μg/mL trypsin). Cell cultures were harvested 24–36 h after infection, at which point all the cells showed a characteristic cytopathic effect. The intracellular viruses were released using three cycles of freeze-thawing. Centrifugation at 4000 g was then performed for 30 min to remove the cell debris. The clarified supernatant was then purified by ultracentrifugation using two concentrations of sucrose solution: 20% (w/w) and 45% (w/w).
-
The ELISAs were conducted as described above. The concentration of the antigens was optimized using a checkerboard titration method. Briefly, various concentrations of antigens were combined with serially diluted serum from highly immunized pigs. The recombinant proteins were diluted in coating buffer at the various concentrations and 100 μL of each recombinant protein was added to the ELISA plates and incubated for 14–16 h at 4 °C. After washing three times with phosphate-buffered saline with Tween 20 (PBST), the plates were blocked with protein-free blocking buffer (Kem-En-Tec Diagnostics, Kuldyssen, Denmark) diluted in PBST for 1 h at room temperature.
The plates were then washed three times with PBST and inoculated with the test samples (100 μL/well). After 1 h incubation at 37 °C, the plates were washed three times with PBST and then inoculated with goat anti-pig IgG antibody labeled with HRP (1:10000). The plates were washed three times with PBST and the substrate, 3, 3′ 5, 5′-tetramethylbenzidine (TMB), was added to each well. Finally, the plates were incubated for 10 min in the dark and then 2mol/L H 2SO 4 was added to terminate the reaction. All the assays were performed in triplicate.
-
The virus neutralization test was performed as previously described (Oh et al., 2005). Briefly, the PEDV GDS01 strain was propagated in Vero cells as described above. All the test sera were inactivated at 56 °C for 30 min and then two-fold serially diluted. The PEDV GDS01 strain at a concentration of 200 50% tissue culture infective doses (TCID 50)/0.1 mL was mixed with an equal volume of diluted sera. The mixture was supplemented with 10 μg/mL trypsin and then incubated for 1 h at 37 °C. Subsequently, Vero cell monolayers on 96-well tissue culture plates were inoculated with 0.1 mL of the mixture. After adsorption for 1 h at 37 °C, the inocula were discarded, and the plates were washed three times with PBS. Maintenance medium containing trypsin (8 μg/mL) was added to each well, and the plates were incubated for 3 days at 37 °C. The SN titers were determined based on the reciprocals of the highest serum dilution that caused inhibition of the cytopathic effect.
-
The correlations between the SN titers and the recombinant S fragments and virus-based ELISA results were evaluated by calculating the correlation coefficients (r).
The OD450 cutoff value for each recombinant S fragment was determined based on the mean ± SD with the PEDV-negative sera. Furthermore, a receiver operating characteristic (ROC) curve analysis was used to test the reliability using the OD450 values obtained from testing the field sow sera. The ROC curves were generated using GraphPad statistical software (GraphPad Software, San Diego, CA, USA). The OD450 cutoff values were selected to provide the optimal sensitivity and specificity for each ELISA. The area under curve (AUC) was used to evaluate the performance of each ELISA.
An amino acid sequence homology analysis was conducted using DNAStar software (version 7, Lasergene Corp., Madison, WI, USA)
-
The post-challenge SN titers for the 156 samples are summarized in Table 1. For the different SN titers, the range of proportions of the 156 samples was 8.3% to 19.2%. More than a quarter (26.4%) of the samples had an SN titer > 1:128. In addition, the results showed that there were 123 positive sera (SN titer > 1:8) and 33 negative sera (SN ≤ 1:8) ( Table 1). Further, the serum samples of different SN titers were used to determine the correlation coefficients of the correlations between the SN titers and ELISA results (involving the recombinant S fragments and the virus antigen). As ROC curves can be used to evaluate the performance of binary classifiers, they have been widely utilized in medical science to determine optimum diagnostic threshold values and to compare the ability of different diagnostic methods to recognize viruses. To determine the reliability of each type of ELISA, a ROC analysis was used for both the positive and negative samples.
SN titer* Number of samples Proportion ≥ 256 21 13.5% (21/156) 128 20 12.9% (20/156) 64 28 17.9% (28/156) 32 24 15.4% (24/156) 16 30 19.2% (30/156) 8 20 12.8% (20/156) ≤ 4 13 8.3% (13/156) Note: * SN titers are expressed as the reciprocal dilution of the serum samples. Table 1. SN titers for 156 sow serum samples from four major swine-rearing provinces in China.
-
The recombinant proteins were expressed and purified after IPTG induction (Figure 2A). Compared with non-induced cell lysate, all the constructs produced antigenic proteins, but they did so to different extents. After purification using Ni-based chromatography, proteins of the predicted sizes were purified (Figure 2B) and the proteins were further confirmed using a western blotting analysis with anti-His-tag antibodies (Figure 2C).

Figure 2. Expression, purification, and identification of the recombinant S protein fragments. (A) Expression of the seven S fragments with or without isopropyl β-D-1-thiogalactopyranoside (IPTG) induction. (B) Recombinant proteins purified using Ni-resin affinity chromatography. (C) Western blotting analysis of the seven recombinant S peptides detected using an anti-His-tag monoclonal antibody.
-
The checkerboard titration method was used to determine the optimum ELISA conditions regarding the coating antigen and detection antibody. The optimum amount of coating antigen was 4.13 μg of recombinant protein per well and the optimum detection antibody dilution ratio was 1:1000. The 123 positive and 33 negative serum samples (as determined based on the SN titers) were used to evaluate the performance of the recombinant S fragment ELISAs.
In addition, the GDS01 virus antigen was prepared as described above. The purified protein band between two concentrations of sucrose was collected with a syringe and used as the ELISA coating antigen. Based on the results of the checkerboard titration method, the optimum dilution ratio for the coating antigen and detection antibody was 1:100 and 1:1000, respectively. The field serum samples were also tested using the virus-based ELISA.
-
The correlation coefficients (r) of the correlation between recombinant S fragment ELISA results and the SN titer were determined, as shown in Figure 3. The r value for SP4 was 0.6113, which is higher than that for SP1, SP2, SP3, SP5, SP6, and SP7 (0.3810, 0.5151, 0.4663, 0.3596, 0.5384, and 0.2660, respectively).

Figure 3. Correlation between the OD450 values (obtained from recombinant S fragment and virus-based ELISAs) and SN titers. Each closed circle represents the OD450 value of a field serum sample plotted against the SN titer. r represents the correlation coefficient
The virus-based ELISA results were also correlated with the SN titer, with an r value of 0.4229. This result was similar to the r value for SP3. In general, when an r value is > 0.4000, the correlation is considered relatively strong. Thus, the correlation was relatively strong between the ELISA results involving SP2, SP3, SP4, SP6, and virus antigen and the SN titer. The r value for SP4 was greater than those for the other recombinant proteins or the virus antigen.
-
The performance of the ELISAs involving the recombinant antigens was analyzed using ROC curves (Figure 4A). The results revealed that the ELISA based on the SP4 antigen (AUC = 0.8538) could differentiate between positive specimens (positive for PEDV neutralizing antibody) and negative specimens (negative for PEDV neutralizing antibody) more precisely than the ELISAs based on the SP1, SP2, SP3, SP5, SP6, and SP7 antigens (AUC = 0.6863, 0.7249, 0.7379, 0.5729, 0.7262, and 0.6085, respectively).
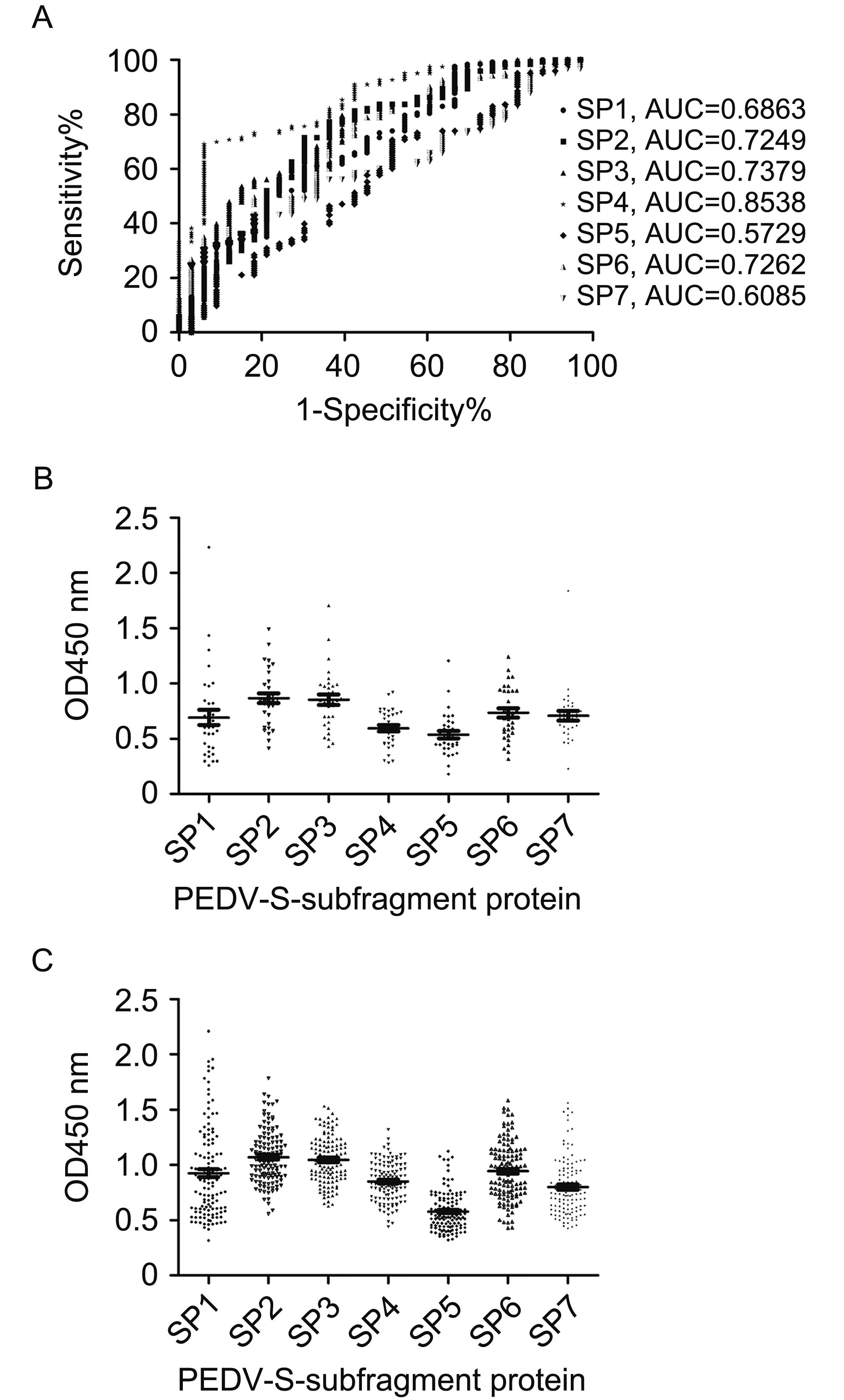
Figure 4. Testing of field serum samples (from sows diagnosed based on SN titers) using recombinant S fragments ELISAs. (A) Graph of the sensitivity versus the false-positive rate (1 - specificity). A ROC analysis was used to determine the discriminatory accuracies of the ELISAs and the AUC was used to evaluate the diagnostic test performance. OD450 values for (B) low-titer PEDV serum samples (n = 33) and (C) high-titer PEDV serum samples (n = 123).
The optimal OD450 cutoff values for the recombinant S fragment ELISAs were determined according to the ROC analyses, as shown in Table 2. Regarding the recombinant S fragment ELISA results, the OD450 values for the negative serum samples (Figure 4B) were lower than those of the positive serum samples (Figure 4C). The sensitivity of each recombinant S fragment ELISA was calculated based on the cutoff value. The ELISA specificities were as follows: SP1: 96.97%; SP2: 96.97%; SP3: 90.91%; SP4: 93.94%; SP5: 90.91%; SP6: 96.97%; and SP7: 96.96%. The ELISA sensitivities were as follows: SP1: 13.01%; SP2: 12.2%; SP3: 39.84%; SP4: 69.11%; SP5: 21.24%; SP6: 26.83%; and SP7: 25.2% (Table 3). Further, the mean OD450 values for the 20 negative sera were as follows: SP1: 0.185; SP2: 0.307; SP3: 0.180; SP4: 0.353; SP5: 0.166; SP6: 0.256; and SP7: 0.206. The sensitivity was 100% when a cutoff value of 0.353 was used for the SP4 ELISA. However, the specificity was too low for this cut-off value to be considered. Therefore, a final cutoff value of 0.774 was selected based on the ROC analysis and samples with OD450 values between 0.353 and 0.774 were considered to be indeterminate.
Recombinant PEDV S proteins PEDV-S-SP1 PEDV-S-SP2 PEDV-S-SP3 PEDV-S-SP4 PEDV-S-SP5 PEDV-S-SP6 PEDV-S-SP7 + – + – + – + – + – + – + – PEDV-positive sow sera (n = 123)a 16 107 15 108 49 74 85 38 26 97 32 91 32 92 PEDV-negative sow sera (n = 33)b 1 32 1 32 3 30 2 31 3 30 1 32 1 32 OD450 cutoff value for ELISA 1.448 1.365 1.106 0.774 0.717 1.123 0.9465 Sensitivity (%)c 13.01 12.2 39.84 69.11 21.24 26.83 25.2 Specificity (%)c 96.97 96.97 90.91 93.94 90.91 96.97 96.97 Note: a 123 serum samples from sows that were diagnosed as PEDV-positive based on the SN titer. The cutoff value was 1:8; b 33 serum samples from sows that were diagnosed as PEDV-negative based on the SN titer. The cutoff value was 1:8;c The sensitivity and specificity for each recombinant protein was calculated based on an OD450 cutoff value determined using a ROC analysis. Table 2. Comparison of the recombinant S fragment ELISA results using 156 field serum samples and SN titers
Samples Mean OD450 ± SD Intra-assay CV*(%) Inter-assay CV*(%) Intra-assay Inter-assay Negative sera 0.206 ± 0.005 0.226 ± 0.014 2.22 6.11 0.308 ± 0.011 0.344 ± 0.038 3.48 11.09 0.279 ± 0.014 0.267 ± 0.045 4.90 16.90 0.381 ± 0.010 0.377 ± 0.047 2.55 12.43 0.280 ± 0.007 0.302 ± 0.018 2.38 5.99 0.250 ± 0.009 0.248 ± 0.017 3.46 6.94 Positive sera 0.838 ± 0.040 0.877 ± 0.050 4.71 5.73 0.673 ± 0.005 0.657 ± 0.047 0.73 7.22 0.760 ± 0.032 0.750 ± 0.040 4.18 5.37 1.065 ± 0.041 1.038 ± 0.115 3.82 11.063 0.938 ± 0.023 0.955 ± 0.051 2.41 5.34 0.548 ± 0.030 0.559 ± 0.047 5.39 8.46 Note: CV*: Coefficient of variation. Table 3. Repeatability of the SP4 ELISA for intra- and inter-assays (with three replications) using neutralizing antibody-negative and -positive serum samples
To assess the newly isolated PEDV virus-based ELISA under field conditions, a panel of the 156 sow serum samples was used. The AUC of the virus-based ELISA was 0.7134. Using a ROC analysis, the cutoff value was 1.233, and the corresponding specificity was 93.94% and the sensitivity was 36.59%. Among the seven S peptides and the virus antigen, the SP4 antigen had the highest sensitivity and specificity.
-
The repeatability of the SP4 ELISA was assessed using six neutralizing antibody-positive samples and six neutralizing antibody-negative samples. All the samples were randomly selected. Three repeats were conducted for each sample for the intra-assays. The repeatability for the inter-assays was also assessed. Finally, the SD and coefficient of variation were calculated to analyze the reliability of the SP4 ELISA. The results revealed that the assay was reliable (Table 3).
Furthermore, we compared the SP4 amino acid sequence of different strains (Supplementary Figure S1). The results showed that the new epidemic strains were highly homologous in terms of the SP4 amino acid sequences. These data suggest SP4 could be used as a promising antigen.
-
For all piglets infected with PEDV, sows are considered to be the source of the infection. Thus, monitoring the sow herd SN status is considered useful for protecting against PEDV. However, the laborious procedure for assessing the SN titer, which requires the production of pure virus, is complicated and expensive. Therefore, recombinant viral proteins expressed in a eukaryotic system have been developed as serological antigens (Gibertoni et al., 2005; Niesters et al., 1986). The PEDV S protein is the only protein for which complementary independent neutralizing epitopes have been identified thus far. The protein is suitable for use in serological testing. However, its high molecular weight (180–220 kDa) (Egberink et al., 1988) makes expression of the full-length recombinant protein difficult.
A full screen of the neutralizing antibody serological testing regions has not yet been carried out on a broad scale. Therefore, in order to clearly map the neutralizing antibody serological testing regions of the entire S1 subunit and the partial S2 subunit, we expressed and identified seven fragments comprising different overlapping regions of the S protein. Among the seven fragments, four were relatively strongly correlated (r value ≥ 0.4) with the SN titer.
The SP7 fragment, which contains two previously identified linear neutralizing epitopes (Hao et al., 2014), had the weakest correlation with the SN titer. The reason behind this may be the amino acid mutations in the epitope or the weak antigenicity of the peptide. The S fragments containing a neutralizing region COE (SP2 and SP6) were relatively strongly correlated with the SN titer, and we speculate that the COE regions may provide a certain level of protection if used in a vaccine, although there have also been amino acid changes in this region (as identified when comparing the current strains to the classic and vaccine strains). Thus, these neutralizing regions (SP2 and SP6) have been considered as candidates for vaccine development (Ge et al., 2012; Oszvald et al., 2007). Overall, the correlation of different recombinant S fragment ELISA results and SN titer showed a highest coefficient of SP4. The correlation coefficient for the correlation between the SP4 ELISA results and SN titers suggests that SP4 is the most promising neutralizing antibody serological testing component.
We also noticed that none of the results showed a high correlation (r ≥ 0.7) between the recombinant S fragment ELISA results and the SN titers. The changing epidemic strains and field sera background factors may have influenced the results (i.e., the serum samples were collected from four provinces and they have otherwise unknown backgrounds, so various factors could have affected the results). Furthermore, different combinations of SP1–SP7 were screened, but no combination provided a better discriminatory result than SP4.
In this study, seven S fragment ELISAs together with the newly isolated GDS01 virus-based ELISA were tested for reliability using ROC analyses. Previous research revealed that trypsin activity in a serum neutralization test might interfere with low dilutions of serum (≤ 1:16) (Oh et al., 2005). In our study, we carefully determined that sera should be categorized as negative for samples with low dilution (≤ 1:8), though this subjective judgment may have led to over- or underestimation. According to the ROC analyses, the AUC for the virus-based ELISA was similar to those of the SP2, SP3, and SP6 ELISAs, but greater than those of the SP1, SP5, and SP7 ELISAs. The SP4 ELISA result revealed that SP4 could be considered as a target serological testing protein, as the AUC was close to 0.9 (indicating that the ELISA could meet the required diagnostic standards) (Huang et al., 2010; Sucheeva et al., 2013).
In this study, a high specificity (> 90%) was the main requirement regarding the optimal OD450 cutoff value of the SP4 ELISA, because the lack of specific herd screening methods can lead to immunity failure in a high-titer herd or insufficient immunity in a low-titer herd. The optimal cutoff value for SP4 indicated high agreement at two SN titer ranges (≥ 1:128 and ≤ 1:8) and lower agreement in the interval between the two ranges (Supplementary Table S2). This indicates that although the serum samples with SN titers of 1:16 to 1:64 cannot be precisely distinguished as positive or negative, the high- and low-titer serum samples could still be distinguished as positive or negative. Thus, the SP4 ELISA can be used to divide the samples to three groups: a high-, low-, and uncertain-titer groups, which could help to determine whether a vaccine candidate is effective.
The disadvantages of prokaryotic expression systems can lead to incorrect protein conformations, but using a baculovirus expression system may solve this problem. Further, we compared the SP4 amino acid sequence of different strains (Supplementary Figure S1). The results showed that the new epidemic strains were highly homologous in terms of the SP4 amino acid sequences. According to research on the IBV S protein, the S2 glycoprotein can induce cross-reactive antibodies and T cell responses that indicate that IBV S2 is relatively conserved (Ignjatovic and Sapats 2005). We speculate that PEDV SP4 has fewer mutations than other fragments as it is located in the S2 domain of the S protein.
In conclusion, effective vaccines and serological testing kits are urgently needed in the field. In our study, we found that the SP4 ELISA result was significantly correlated with the SN titer. In addition, the ROC analysis indicated that the SP4 ELISA is suitable for herd infection status testing. Moreover, the SP4 amino acid sequence of different strains (including vaccine strains) is highly conserved, which indicates that this recombinant protein is a promising antigen for use in PEDV control strategies.
-
This work was supported by the National Key Research and Development Program (2016YFD0500101).
-
The authors declare that they have no conflict of interest. The animal study was supervised by the Institutional Animal Care and Use Committee of the Sun Yat-sen University (IACUC DD-17-0403) and used in accordance with regulation and guidelines of this committee.
-
JH and YZ designed the experiments. JH, YZ, ZW, and XZ carried out the experiments. SF analyzed the data. JH and YZ wrote the manuscript. YC supervised the study and finalized the manuscript. All authors read and approved the final manuscript. Supplementary figure /tables is available on the websites of Virologica Sinica: www.virosin.org; link.springer.com/journal/12250.
-

Table S1. Primers used for polymerase chain reaction (PCR) involving the seven recombinant S peptides

Figure S1. Alignment of the deduced amino acid sequence of the partial S2 subunit. Amino acid residues 791–954 of the S2 subunit of the PEDV GDS01 strain were aligned with those of other PEDV strains (including vaccine strains, which are marked with asterisks, and newly isolated strains from various areas). The dots indicate the regions where the sequences are identical to the consensus sequence

Table S2. Comparison of SP4 ELISA results with SN titers based on testing 156 field sow serum samples
Evaluation of purified recombinant spike fragments for assessment of the presence of serum neutralizing antibodies against a variant strain of porcine epidemic diarrhea virus
- Received Date: 06 March 2017
- Accepted Date: 14 June 2017
- Published Date: 20 July 2017
Abstract: Since 2010,variant strains of porcine epidemic diarrhea virus (PEDV) have caused disasters in the pork industry.The spike (S) protein,as the major immunity-eliciting antigen,has previously been used for serological testing and has been found to correlate significantly with the results of the serum neutralization (SN) test.However,further evaluation of this method is needed as new epidemic strains of PEDV emerge.Hence,the main objective of this study was to assess sow sera and determine the correlation between enzyme-linked immunosorbent assay (ELISA) results (involving a newly isolated GDS01 virus-based ELISA and ELISAs based on seven recombinant fragments comprising overlapping S1 and partial S2 sequences) and SN titers.Furthermore,we determined the reliability of the ELISAs based on receiver operating characteristics (ROC) curve analyses.For the most promising ELISA,i.e.,the SP4 ELISA,the correlation coefficient (r) and the area under curve (AUC) were determined to be 0.6113 and 0.8538,respectively.In addition,we analyzed the homology of the SP4 sequences obtained from different strains (including vaccine strains) and found that various strains showed a high degree of homology in this region.Thus,we conclude that SP4 is a promising serological testing protein for use in the field.

















 DownLoad:
DownLoad: