HTML
-
Porcine reproductive and respiratory syndrome (PRRS) is an economically important swine disease characterized by severe reproductive failure in sows and respiratory distress in piglets and growing pigs and represents a major threat to the worldwide swine industry (Corzo et al. 2010; Han et al. 2017). It is estimated to cost the pork producers at least $600 million annually in North America with comparative losses in most other countries (Neumann et al. 2005). PRRS virus (PRRSV), the causative agent, is an enveloped, single-stranded positive-sense RNA virus and belongs to the newly classified genus Porartevirus of the family Arteriviridae in the order Nidovirales (Kuhn et al. 2016). In 2006, a highly pathogenic PRRSV (HP-PRRSV) of type 2, the etiological agent of unknown swine "high fever" disease, broke out in China and surrounding nations and has been plaguing the Chinese swine industry (Tian et al. 2007; Zhou and Yang 2010). Since 2013, several field PRRSV isolates in China have been found to share high nucleotide similarity to a group of strains represented by NADC30, a type 2 PRRSV that was previously reported in USA in 2008 (Brockmeier et al. 2012; Zhou et al. 2015; Li et al. 2016). Although the virulence of NADC30-like strains is relatively mild, they are now starting to recombine with the HP-PRRSV, making the PRRSV control in China more complex (Han et al. 2017; Tian 2017).
The PRRSV genome is approximately 15 kb long and contains at least 12 known open reading frames (ORFs)—ORF1a, ORF1b, ORFs2 (2a and 2b), ORF3, ORF4, ORF5 (5a and 5), ORF6 and ORF7 (Lunney et al. 2016). Among the ORFs, ORF1a and ORF1b occupy the 5' terminal twothird of the genome and specify the viral replicase proteins. Translation of ORF1a yields the replicase polyprotein 1a (pp1a) whereas ORF1b is expressed through -1-programmed ribosomal frameshifting (-1PRF) mechanism in the ORF1a/ORF1b overlapping region, which C-terminally extends pp1a into pp1ab with an efficiency of about 10%– 20% (den Boon et al. 1991; Snijder et al. 2013). PPRSV pp1a is processed into at least 12 mature nonstructural proteins (nsps), including nsp1α, nsp1β, nsp2, nsp2TF, nsp2 N, nsp3, nsp4, nsp5, nsp6, nsp7α, nsp7β, and nsp8, while pp1b produces the additional proteins that are nsp9–12, which have homologs across arterivirus family (Snijder et al. 1994; den Boon et al. 1995; van Dinten et al. 1996; Wassenaar et al. 1997; Gorbalenya et al. 2006; Fang et al. 2012). The proteolytic maturation is thought to be mediated by virally encoded proteases located within nsp1, nsp2 and nsp4. The papain-like protease domain (PLP2) within nsp2, which has both tans- and cis-cleavage activities, is responsible for the cleavage at the nsp2–3 junction (Snijder et al. 1995; Han et al. 2009) while the remaining cleavages in both pp1a and pp1ab are likely carried out by the 3C-like main protease within nsp4 (Snijder et al. 1996; Tian et al. 2009).
The nsp9 replicase protein is a critical determinant for the fatal virulence of the Chinese HP-PRRSV (Li et al. 2014; Xu et al. 2018; Zhao et al. 2018). This protein specifies the putative function of viral RNA-dependent RNA polymerase (RdRp) and is the major engine for catalyzing the viral RNA synthesis during PRRSV infection (van Dinten et al. 1996; Beerens et al. 2007; Posthuma et al. 2017). Comparative bioinformatics analyses based on the studies of equine arteritis virus (EAV) and other positive-stranded RNA viruses have localized the viral RdRp domain to the C-terminal half of nsp9 (Gorbalenya et al. 1989) and assigned nidovirus RdRp-associated nucleotidyltransferase domain (NiRAN) to the N-terminal region of ORF1b-encoded portion (Lehmann et al. 2015). As a result of the frameshift strategy, nsp9, the first protein within ORF1b, is expressed during infection as the C-terminal extension of nsp8, a small viral nsp that is encoded by ORF1a (Fang and Snijder 2010; Posthuma et al. 2017). However, it has remained unclear whether the mature form of nsp9 in virus-infected cells still retains nsp8. Addressing this question is clearly critical to understand the biological function of nsp9. By taking advantage of specific polyclonal antibodies to both nsp8 and nsp9, here we analyzed the relationship between nsp8 and ORF1b-encoded nsp9 in PRRSV-infected cells. Our studies provide evidence to suggest that nsp8 serves as the N-terminal portion of nsp9.
-
Both human embryonic kidney (HEK) 293FT cells and MARC-145 cells were maintained in GIBCO Dulbecco's modified Eagle medium DMEM (Fisher Scientific, Waltham, MA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS) (HyClone Laboratories Inc., South Logan, UT). Porcine pulmonary alveolar macrophages (PAMs) were prepared as previously described (Zhang et al. 2009) and maintained in RPMI-1640 (Fisher Scientific) supplemented with 10% (v/v) FBS. The Chinese highly pathogenic PRRSV strain JXwn06 (GenBank accession no. EF641008.1) used in this study has been described previously (Zhou et al. 2009). Infected MARC- 145 cells or PAMs were maintained in DMEM or RPMI- 1640 medium supplemented with 2% (v/v) FBS. All the cells were maintained in a humidified incubator with 5% CO2 at 37 ℃.
-
All plasmids were constructed by standard recombinant DNA techniques. Briefly, the nsp8 and nsp9 genes of PRRSV strain JXwn06 were amplified by PCR and then cloned into vector pGEX-6p-1 and pEGFP-N2 (Clontech, Palo Alto, CA, USA) to generate plasmids pGST-nsp8, pHA-nsp8, and pNsp9ORF1b. The nsp8–9ORF1b gene of PRRSV strain JXwn06 was cloned by PCR, and the ORF1a/1b ribosomal frameshift site in the nsp9 gene was removed by overlapping PCR as described before (Beerens et al. 2007; Liu et al. 2015), and then cloned into vector pEGFP-N2 to generate pNsp8–9ORF1b. The primers used for plasmid construction were listed in Supplementary Table S1. All recombinant plasmids were further verified by DNA sequencing. The pHA-nsp4, pFlag-nsp4 and pFlag-Cyto.c1 have been described previously (Zhang et al. 2017b). The plasmid pHA-nsp2/3ΔPLP2 was constructed by cloning the fragment coding for nsp2–3 but excluding the PLP2 coding sequence into pEGFP-N2. The plasmid pHA-nsp2/3ΔPLP2 G1166P was constructed by introducing a point mutation (G1166P) at the nsp2/3 cleavage site. The plasmid pMyc-PLP2 made by cloning the coding sequence for nsp2 PLP2 (nsp2 aa. 12-323) into pEGFP-N2.
-
The plasmid pGST-nsp8 was transfected into E. coli BL21 (DE3) competent cells (TransGens Biotech, Beijing, China). Cultures in logarithmic phase (at OD600 of ~ 0.6–0.7) were induced at 30 ℃ for 6 h by adding isopropyl-β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich) to the medium at a final concentration of 0.5 mmol/L, followed by centrifugation for harvest. The pellets of 150 mL cultured bacteria were washed twice by using 1 × phosphate-buffered saline (PBS) and then resuspended with 30 mL PBS. The bacterial cells were lysed by sonication, after which the soluble (supernatant) and insoluble (inclusion body) fractions of GST-nsp8 were separated by centrifugation at 12, 000 rpm, 4 ℃ for 20 min. The abundance of GST-nsp8 in the two fractions was analyzed by SDS-PAGE. Having known the soluble fraction contains the majority of GST-nsp8, the Glutathione SepharoseTM 4B beads (GE Healthcare Bio-Science, Chicago, IL, USA) were used to purify the recombinant GST-nsp8 protein from the supernatant, according to the manufacturer's instructions. The concentration of purified GST-nsp8 protein was measured by using Ultra-trace nucleic acids and protein detector (Thermo Fisher Scientific, Waltham, MA, USA). The purity and antigenicity of purified GST-nsp8 protein were analyzed by SDS-PAGE and Western blot analysis, respectively.
-
The GST-nsp8 specific polyclonal antibodies (pAbs) were generated by following the previously established protocols (Bao et al. 2015), but with a few modifications. Briefly, 3-month-old (about 2 kg) New Zealand white rabbits were injected subcutaneously with 1.0 mg per kg (body weight) purified GST-nsp8 protein emulsified with an equal volume of Freund's complete adjuvant (Sigma-Aldrich). After 2 weeks, the rabbits were immunized again with 0.5 mg per kg (body weight) purified GST-nsp8 protein emulsified with an equal volume of Freund's incomplete adjuvant (Sigma-Aldrich). The third and fourth immunizations were executed in the same manner as the second one at 2-week intervals. Blood of rabbits was collected 7 days after the fourth immunization, and the serum was separated following a centrifugation of 5000 rpm for 10 min after stayed in 37 ℃ for 2 h.
PRRSV nsp7 were expressed as a fusion protein with GST in E. coli BL21 cells and the purified recombinant GST-nsp7 was used as the immunogen. The nsp9 antibodies were made by using ORF1b-coded nsp9 tagged with strepII as an immunogen that was expressed and purified from E.coli BL21 cells. The immunization protocol was the same as that for nsp8. Reactivity and specificity of the respective antibodies were tested in the condition of transfection and infection meanwhile the mock transfected or mock-infected cells serve as negative control. The normal rabbit serum was also used as a negative control. All the antibodies showed good reactivity and specificity.
The mouse anti-HA (H3663) and anti-Flag (F1804) monoclonal antibodies (mAbs) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Mouse anti-GST Tag (12 GB) mAb was purchase from Abmart (Shanghai, China). Mouse anti-nsp9, anti-nsp10, and anti-nsp4 mAbs were prepared in our laboratory (Zhang et al. 2017b). The Alexa-fluor-488-conjugated goat anti-rabbit IgG(H + L) F (ab')2 fragment, alexa-fluor-568-conjugated goat anti-rabbit IgG(H + L) F (ab')2 fragment, alexa-fluor-488-conjugated goat anti-mouse IgG(H + L) F (ab')2 fragment and Alexa-fluor-568-conjugated goat anti-mouse IgG(H + L) F (ab')2 fragment were all purchased from Invitrogen (Carlsbad, CA, USA). Hoechst 33258 (C1011) was purchased from Beyotime Biotechnology (Nantong, China).
-
For Western blot, the cells seeded in plates were washed three times with ice-cold 1 × PBS, then lysed in ice-cold lysis buffer [50 mmol/L Tris–HCl (pH 7.4), 1.0 mmol/L EDTA, 150 mmol/L NaCl, 5 mmol/L MgCl2, 10% Glycerol and 1% TritonX-100] supplemented with 1 × protease inhibitor cocktail (Sigma) for 30 min with gentle rotation. Following centrifugation at 12, 000 × g for 30 min at 4 ℃, the supernatant was transferred to a fresh tube. The cell lysates were boiled in 1 × loading buffer for 10 min and resolved by SDS-PAGE with 8% or 15% polyacrylamide gels. The proteins were electrically transferred onto a 0.22 lm polyvinylidene fluoride membrane (PVDF) (Mick Millipore, Darmstadt, Germany). The membrane was blocked in 5% skim milk powder in PBS for 1 h at room temperature (RT) and probed with indicated primary antibodies diluted in the primary antibody diluents (TOYOBO, Japan) for 2 to 4 h at RT. After being washed three times by PBST (PBS with 0.5% Tween-20), the membrane was subsequently hybridized with either appropriate horseradish peroxidase (HRP)-conjugated goat anti-rabbit (ZB-2301) or goat anti-mouse (ZB-2305) (ZSGB-BIO, Beijing, China) secondary antibody at a dilution of 1:10, 000 and incubated at RT for 1 h. After being washed three times by PBST, the membrane was probed by the ECL Western blot system (Invitrogen) and exposed to a FluorChem E apparatus (ProteinSimple, Santa Clara, CA, USA).
-
MARC-145 cells grown on coverslips in 6-well plates were either transfected with relevant plasmids or infected with PRRSV strain JXwn06 at a multiplicity of infection (MOI) of 0.1. At 24 h post transfection or infection, the cells were fixed with 4% formaldehyde for 8 min, permeabilized with 0.3% Triton X-100 in PBS (pH 7.2), and blocked with 2% bovine serum albumin (BSA). Cells were incubated with appropriate primary antibodies for 1 h at RT, and then washed three times with 1 × PBS. After that, the cells were incubated with appropriate secondary antibodies. Nuclear DNA was stained by Hoechst (Beyotime Biotechnology, China) for 10 min at RT. The images were captured under an Olympus confocal microscope (Fluoview1000) and processed by using Image J.
-
MARC-145 cells grown on T75 flasks were infected with PRRSV JXwn06 at a MOI of 1. At 24 h post infection, the cell lysates were prepared as described above in the "Western blot analysis" section and then precleared with protein A/G agarose beads (Santa Cruz, CA, USA). The nsp8 or nsp9 in the supernatant was then immunoprecipitated using antibodies to nsp9 or nsp8 in conjunction with protein A/G Sepharose beads (GE Healthcare Bio-Science) at 4 ℃ overnight on a rocker. The beads-protein complexes were collected by a centrifugation of 4, 000 rpm for 2 min at 4 ℃ and gently washed 8 times with ice-cold IP buffer [50 mmol/L Tris–HCl (pH 7.4), 1.0 mmol/L EDTA, 150 mmol/L NaCl, 5 mmol/L MgCl2, 10% Glycerol and 0.5% TritonX-100] supplemented with 1 × protease inhibitor cocktail and once with PBS. The immunoprecipitated proteins were then subject to SDS-PAGE and Western blot analysis.
Cell, Virus and Infection
Plasmid Construction
Prokaryotic Expression and Purification of Recombinant PRRSV nsp8 Protein
Antibodies and Chemicals
Western Blot Analysis
Confocal Immunofluorescence Assay
Co-immunoprecipitation (co-IP) Assay
-
The replicase protein nsp8 is highly conserved among various PRRSV strains and has a size of 45 amino acids (Music and Gagnon 2010). To facilitate the production of antibodies to PRRSV nsp8, the gene coding for nsp8 was cloned from HP-PRRSV strain JXwn06 and expressed as a GST fusion protein in E. coli BL21 (DE3) cells (Fig. 1A). After ultra-sonication, the recombinant protein was detected predominantly in the supernatant with very little in pellet (Fig. 1A, lane 3 and 4), indicating that GST-nsp8 mainly exists as a soluble form instead of inclusion bodies. The GST-nsp8 protein (soluble form) was subsequently purified by Glutathione SepharoseTM 4B beads to homogeneity (Fig. 1A, lane 5), and the identity was further confirmed by Western blot analysis with anti-GST monoclonal antibodies as shown in Fig. 1B (lane 3).
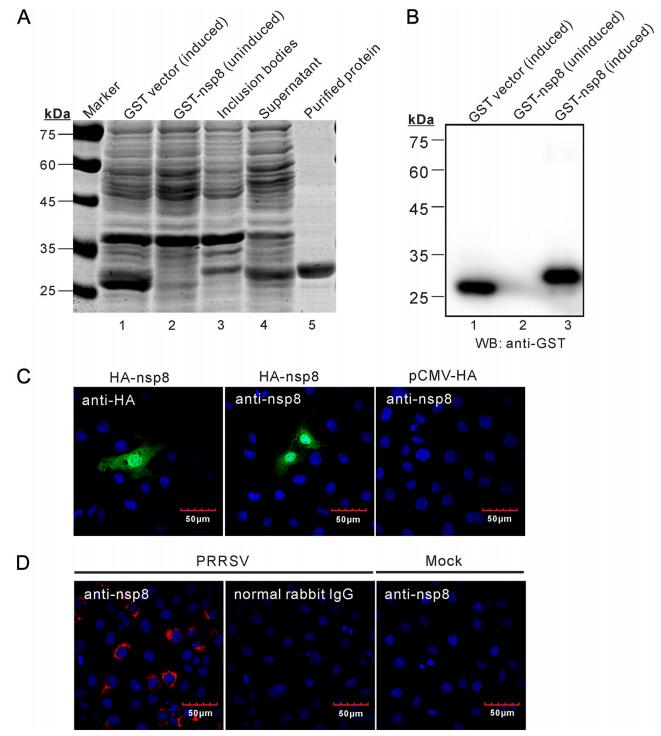
Figure 1. Preparation of rabbit polyclonal antibodies to PRRSV nsp8. (A) Analysis of the expression and purification of recombinant GSTnsp8 protein by 12% SDS-PAGE gel. (B) Western blot of the expression of GST-nsp8 by anti-GST mAb. (C) Immunofluorescence detection of HA-nsp8 in transfected MARC-145 cells by anti-GSTnsp8 serum. MARC-145 cells on coverslips in six well plates were transfected with the plasmid pHA-nsp8 (middle) or the vector pCMVHA (right). At 18–24 h post transfection, the cells were fixed, permeablized and stained with proper antibodies to the HA epitope or to GST-nsp8, followed by Alexa Fluor 488-conjugated secondary antibodies. The cell nuclei were stained with Hoechst (blue) and examined by confocal microscopy. (D) Detection of nsp8 expression in PRRSV infected MARC-145 cells. MARC-145 cells grown on coverslips were either mock infected with DMEM or infected with HP-PRRSV strain JXwn06 at an MOI of 0.1. At 24 h after infection, the cells were fixed and stained with rabbit pre-immune serum or antiGST-nsp8 serum and examined by confocal microscopy.
The purified GST-nsp8 was then used as the immunogen to make polyclonal antibodies in rabbits. The pre-immune and immune sera were collected as negative IgG control and anti-nsp8 pAb, respectively. To test the specificity of the anti-nsp8 polyclonal antibodies, HA-nsp8 was transiently expressed in MARC-145 cells. At 24 h post transfection, its expression was analyzed by immunofluorescence assay (IFA) with antibodies to GSTnsp8 or to the HA epitope tag (Fig. 1C). The results showed that HA-nsp8 was mostly localized to nucleus but with a diffusion pattern in the cytoplasm as inferred by the staining with anti-HA antibodies. The antibodies to GSTnsp8 revealed similar localization pattern of nsp8. The specificity was further confirmed by staining the cells transfected with the plasmid pCMV-HA, which gave no detectable signals. We also investigated the expression of nsp8 under the condition of infection. As show in Fig. 1D, the staining with GST-nsp8 antibodies gave positive signals around perinuclear region in PRRSV-infected but not in mock-infected MARC-145 cells. In addition, the preimmune rabbit serum staining gave no detectable signal in PRRSV-infected cells. Together, we conclude that the GST-nsp8 immune serum contains antibodies specific to PRRSV nsp8 and that the prepared anti-nsp8 polyclonal antibodies possess good specificity.
-
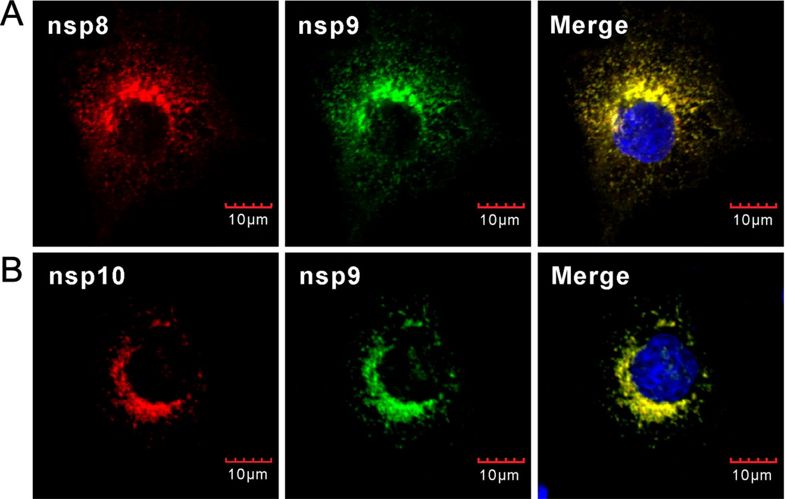
The perinuclear distribution of nsp8 in virus infected cells is consistent with that of the previous report by using type Ⅰ PRRSV (Li et al. 2012). To analyze the relationship between nsp9 and nsp8, we investigated whether nsp9 colocalizes with nsp8 in PRRSV-infected MARC-145 cells. MARC-145 cells were infected with PRRSV strain JXwn06 at a MOI of 0.1, and at 24 h post infection, IFA was performed with nsp9 probed with monoclonal antibody recognizing the region located within ORF1b-encoded portion. As shown in Fig. 2A, nsp9 showed a perfect colocalization relationship with nsp8 in cytoplasm. As a control, the RNA helicase protein nsp10 also well co-localized with nsp9 in cytoplasm in PRRSV-infected MARC-145 cells (Fig. 2B). Thus, the results suggest that nsp8 or nsp8-containing nsp intermediates were recruited to the viral replication site during infection.
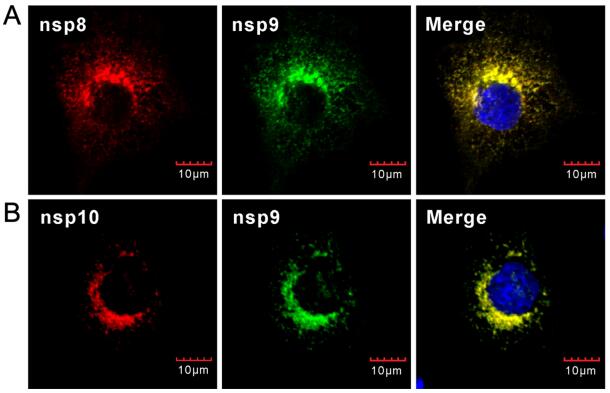
Figure 2. Co-localization analysis of nsp9 with nsp8 and nsp10 in PRRSV-infected cells. MARC- 145 cells infected with PRRSV strain JXwn06 at a MOI of 0.1 were fixed and double stained at 24 h post infection with antinsp9 monoclonal antibodies (mAb) and anti-nsp8 polyclonal antibodies (pAb) (A), or with anti-nsp9 pAb and anti-nsp10 mAb (B). Nuclei were stained with Hoechst. Representative images were taken by a confocal microscope.
-
We next probed the relationship between nsp8 and nsp9. In PRRSV MARC-145 cells infected cells, both anti-nsp9 mAb and rabbit polyclonal antibodies (pAb) recognized a major band with a mobility between 72 and 95 kDa (Fig. 3A, lane 2). The molecular weight of this band was much larger than the predicted size of ORF1b-coded nsp9, which contains 640 amino acids with a size of about 70 kDa (Supplementary Table S2) (Music and Gagnon 2010). In addition, several very minor bands (26–34 kDa, 43–55 kDa, 55–72 kDa of nsp9 could sometimes be discerned (Fig. 3A, lane 2 and Fig. 3C, lane 2). The nature of these bands was not clear and could represent nonspecifically degraded products during sample preparation. For nsp8, it is a 45-amino-acid protein and the detection of this small molecule is beyond the resolution of this 10%–15% gel system. Interestingly, the polyclonal antibodies to nsp8 detected three distinct species with apparent different mobilities in the gel (Fig. 3B, lane 2). The slowest moving species had a size similar to the 72–95 kDa band recognized by anti-nsp9 antibodies (Figs. 3A, 4B, 4C). The other two species corresponded in size to the processing intermediates nsp5–8 and nsp7–8. In the co-immunoprecipitation assay (Co-IP), we found that anti-nsp8 antibodies could pull down the 72–95 kDa product recognized by anti-nsp9 antibodies (Fig. 3C). However, the minor bands that were recognized by nsp9 antibodies (Fig. 3A, lane 2 and Fig. 3C, lane 2) were not pulled down by antibodies to nsp8 (Fig. 3C, lane 5), suggesting that the minor bands represent the portions of ORF1b-coded nsp9). More significantly, the nsp8 antibodies could detect the 72–95 kDa band pulled down by nsp9 antibodies (Fig. 3D, lane 5). Thus, these results indicated that both anti-nsp9 mAb and anti-nsp8 pAb can specifically recognize the same 72–95 kDa protein in PRRSV-infected MARC-145 cells.
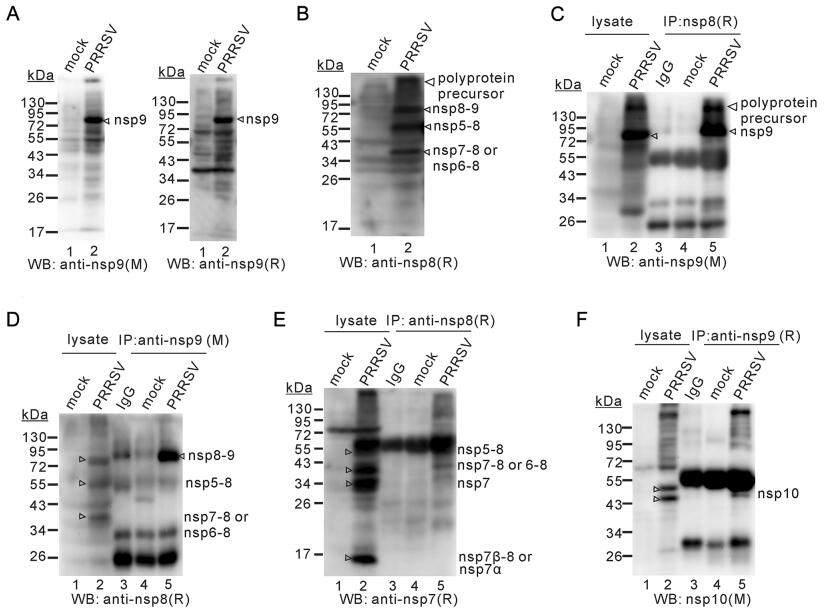
Figure 3. Nsp8 serves as the N-terminus of nsp9. MARC-145 cells were either mock-infected or infected with PRRSV strain JXwn06 at a MOI of 1 for 24 h, and the cell lysates were subjected to Western blot analysis. (A) Detection of nsp9 product(s) by anti-nsp9 monoclonal antibodies (mAb, left) and polyclonal antibodies (pAb, right). (B) Detection of nsp8 products by anti-nsp8 pAb. (C and D) Co-IP analyses by using antibodies to nsp9 and nsp8. (E) Co-IP analysis of nsp8 products by anti-nsp7 pAb. (F) Co-IP analysis of nsp9 products by anti-nsp10 mAb. M: mouse monoclonal antibodies; R, rabbit polyclonal antibodies.
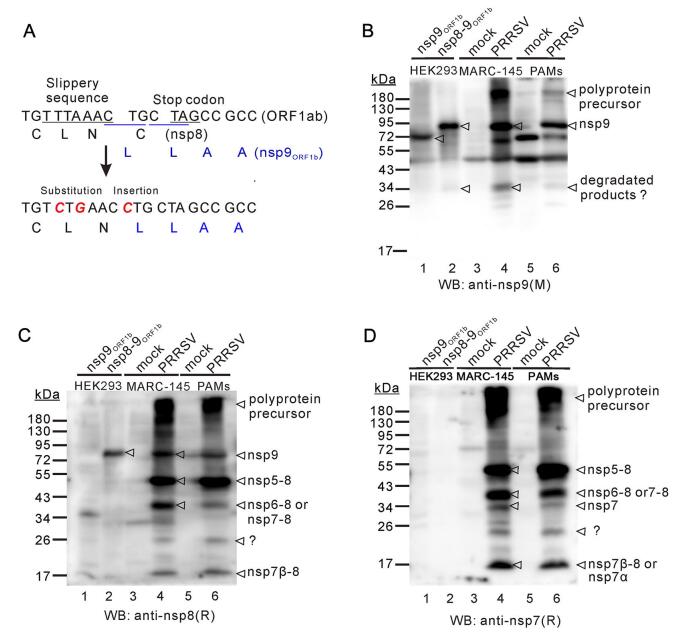
Figure 4. The in vitro expressed Nsp8–9 has the similar size as the native nsp9. (A) Mutagenesis of nsp8–9 junction site to allow inframe expression of nsp9. (B, C and D) Western blot analysis of nsp9, nsp8–9 expressed in HEK 293FT cells, and endogenous nsp9 from PRRSV JXwn06-infected MARC-145 cells and PAMs by using antinsp9 monoclonal antibodies (mAb) (B) or anti-nsp8 polyclonal antibodies (pAb) (C) or anti-nsp7 pAb (D) as primary antibodies. M: mouse monoclonal antibodies; R, rabbit polyclonal antibodies.
To test whether nsp7 or nsp10 is part of the 72–95 kDa product, we used antibodies to nsp7 and nsp10 for further investigation (Fig. 3E, 3F). We found that nsp7 antibodies recognized mainly 4 different species (Fig. 3E, lane 2), corresponding to nsp5–8, nsp7–8, nsp7 and nsp7a, as being judged by the mobility of the moving species. Consistent with previous report (Zhang et al. 2018), antibodies to nsp10 detected two major bands with a molecular size ranging from 43 to 55 kDa, corresponding to the two isoforms of nsp10 (Fig. 3F, lane 2). However, both antibodies failed to detect the 72–95 kDa product (Fig. 3E, 3F). In the Co-IP assay, nsp7 antibodies did not recognize the 72–95 kDa band pulled down by nsp8 antibodies (Fig. 3E), and nsp10 antibodies also failed to detect the 72–95 kDa product pulldown by anti-nsp9 antibodies (Fig. 3F), suggesting that the 72–95 kDa product does not contain either nsp7 or nsp10. Taken together, we conclude that the predominant form of nsp9 contains nsp8 but not nsp7 or nsp10.
-
To further analyze the size of the 72–95 kDa nsp9, we transiently expressed nsp8–9 and the ORF1b-coded nsp9 (nsp9ORF1b) in HEK 293FT cells. To express the nsp8–nsp9 fusion protein (nsp8–9ORF1b), nsp9 was made in frame with nsp8 by inactivation of the frameshift signal (Fig. 4A). Meanwhile, MARC-145 cells and PAMs were infected with PRRSV JXwn06 at an MOI of 1, and at 24 h post infection, the cells were harvested and whole cell lysates were prepared. All the prepared cell lysate samples were subjected to Western blot analysis and reacted with antibodies to nsp8 and nsp9. The results showed that the expression pattern of nsp9 in infected PAMs was very similar to that in MARC-145 cells and the major band of nsp9 had the similar size in both cell types (Figs. 3A-3C, 4B) and that nsp8–9ORF1b, but not nsp9ORF1b, had the similar mobility as the 72–95 kDa nsp9 (Fig. 4B, 4C). In addition, similar expression pattern for nsp7 and nsp8 products or intermediates could be discerned in both cell types (Fig. 4C, 4D). Overall, these results provide further evidence to support the notion that nsp8 is part of nsp9.
-
To detect whether nsp8–9 could be cleaved by PLP2 or nsp4, EGFP-nsp8–9-HA was co-expressed with Myc-PLP2 or Flag-nsp4 in HEK 293FT cells. At 36–48 h post transfection, the cells were harvested and subjected to Western blot for cleavage analysis. As shown in Fig. 5A, PRRSV PLP2 did not cleave EGFP-nsp8–9-HA (Fig. 5A, line 2). In contrast, it cleaved readily the substrate HA-nsp2/3ΔPLP2 (Fig. 5A, lane 4). When the cleavage site G|G dipeptide was mutated, PLP2 failed to cleave HA-nsp2/3ΔPLP2 G1166P (Fig. 5A, lane 5). For the nsp4 cleavage assay, the EGFP-nsp8–9-HA could not be cleaved by nsp4 either (Fig. 5B, lane 1), while Flag-cytochrome c1 (Flag-Cytoc1) could be cleaved by nsp4 (Fig. 5B, line 3) as previously reported (Zhang et al. 2017a). These results were also consistent with the bioinformatics analysis which does not predict cleavage sites for nsp4 or PLP2 at the nsp8–9 junction site. Together, these results suggest that nsp8–9 does not appear to contain the nsp2 or nsp4 cleavage site.
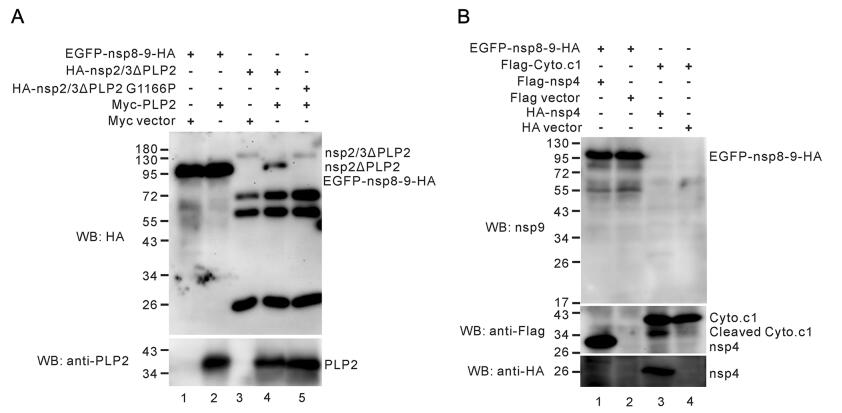
Figure 5. Neither nsp4 nor PLP2 cleaves nsp8–9 in transfected cells. (A) HEK 293FT cells were transfected to express EGFP-nsp8–9-HA or HA-nsp2/3ΔPLP2 or HA-nsp2/3ΔPLP2 G1166P, together with or without Myc-PLP2. At 36–48 h post transfection. The HA-nsp2/ 3ΔPLP2 and HA-nsp2/3ΔPLP2 G1166P were precipitated by anti-HA monoclonal antibodies (mAb) in conjunction with protein A/G Sepharose beads. The cleavage of the substrates was analyzed by Western blot with anti-HA mAb and expression of PLP2 was detected anti-nsp2-PLP2 polyclonal antibodies (pAb). (B) HEK 293FT cells were transfected to express EGFP-nsp8–9-HA together with or without Flag-nsp4, and to express Flag-Cyto.c1 together with or without HA-nsp4. At 36–48 h post transfection, the cleavage of the substrates was analyzed by Western blot with antibodies to nsp9, Flag and HA epitopes.
Production of Rabbit Polyclonal Antibodies to PRRSV Nsp8
Nsp9 Co-Localizes with Nsp8 in PRRSV-Infected Cells
Nsp9 Exists as a Predominant 72–95 KDa Product in PRRSV-Infected Cells and Contains Nsp8 at Its N-Terminus
In Vitro Expressed Nsp8–9 has the Similar Mobility as the Native Nsp9 in PRRSV-Infected Cells
Either PLP2 or Nsp4 Does Not Cleave Nsp8–9 in Transfected Cells
-
The PRRSV nsp9 encodes the function of viral RdRp and is the engine driving viral RNA synthesis (van Dinten et al. 1996; Beerens et al. 2007; Posthuma et al. 2017); it is encoded within ORF1b and translated through the C-terminal extension of nsp8 during infection (Meulenberg et al. 1993; Kappes and Faaberg 2015). However, it is not clear whether the mature form of nsp9 retains nsp8, although the nsp8–9 does not contain the predictable cleavage sites for nsp2 or nsp4. Our studies here identified nsp9 as a major product with a size between 72 and 95 kDa. Also, we provided three pieces of evidence on nsp8 being part of nsp9 as follows. (ⅰ) The in vitro expressed nsp8–9, but not the ORF1b-coded portion of nsp9, had the similar mobility on the gel as the 72–95 KDa form of nsp9 from virusinfected cells; (ⅱ) neither nsp4 nor nsp2 PLP2 cleaves nsp8–9 in vitro; and (ⅲ) the nsp8 antibodies, but not the antibodies to nsp7 and nsp10, could detect a major product that had the similar size to the native nsp9. Furthermore, nsp9 could be co-immunoprecipitated by antibodies to nsp8, and vice versa. These findings here should pave the way for further charactering the biological function of PRRSV nsp9.
The nsp8–9 protein of PRRSV strain JXwn06 contains 685 amino acids and has a predicted size of around 75 kDa. In the transfected cells, the ORF1b-coded nsp9 had a size of around 70 kDa. However, fusion expression of nsp8 and nsp9 increased the size dramatically (Fig. 4B), and the difference appeared much larger than the size of nsp8 itself (4.8 kDa) (Fig. 4B). These results suggest that nsp8 may exert an unknown effect on the structure of ORF1b-encoded nsp9 or affect its post-translational modifications, or both, leading to a change in the mobility of nsp9 in the gel. In the virus infected cells, the predominant product recognized by nsp9 antibodies had a molecular weight between 72 kDa and 95 kDa, similar to the size of the in vitro expressed nsp8–9. More over, this product does not contain nsp10 or nsp7, which is evidenced by the direct western blot analysis of PRRSV-infected cell lysates with antibodies to nsp10 and nsp7 (Fig. 3E, 3F). One caveat from these studies is that we do not know if the nsp9 product contains the intact nsp8. This needs to be further clarified in the near future.
Our studies also provide insights into the maturation process of PRRSV replicase polyproteins. The polyproteins pp1a and pp1ab can be proteolytically cleaved via the major and minor pathways (Ziebuhr et al. 2000; Li et al. 2012, 2015). In addition to generation of the individually mature nsps, intermediate processing products have also been observed in PRRSV-infected cells (Li et al. 2015). By using type Ⅰ PRRSV as the model organism, Li et al. (2012) detected the presence of the long-lived processing intermediates, such as nsp3–4, nsp5–7, nsp5–8 and nsp3–8, which likely play an important role in the virus life cycle. In this study, we were also able to detect the presence of nsp intermediates. There are two major products that could be detected by antibodies to both nsp7 and nsp8 (Fig. 3B, 3E). One product had the size of around 55 kDa, and this may correspond to the nsp5–8 intermediate (490 aa, Supplementary Table S2), which has a predicted molecular weight of 54.3 kDa. The other band had a mobility between 34 kDa and 43 kDa and corresponds to the intermediate nsp7–8 (304 aa) with predicted size of 33.7 kDa, or nsp6–8 (320 aa) with predicted size of 35.4 kDa. The 34 kDa species detected by nsp7 antibodies may represent the unprocessed full-length nsp7 (259 aa) or nsp6–7 (275 aa), whereas the 17 kDa protein may represent the processed nsp7α. Thus, the nsp5–8, nsp7 and nsp7–8/6–8 appear to be the major long-lived intermediates in HPPRRSV infected cells. The biological functions of these proteins await to be explored in the future.
-
This work was supported by the National Key Basic Research Plan Grant from the Chinese Ministry of Science and Technology (2014CB542700), the China National Thousand Youth Talents program (1051-21986001), and the earmarked fund for China Agriculture Research System (CARS-35) from the Chinese Ministry of Agriculture.
-
HY, JH and YL conceptualized and designed the study. YL and JS performed the experiments in the study. YH, YC, LPL, JS, SZ and JS contributed reagents to this study. JH and YL analyzed the data. ZL, XNG and XG contributed to the study design. JH, YL and HY wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Author Contributions
-
The authors declare that they have no conflict of interest.
-
The animal experiments in this study were approved by The Laboratory Animal Ethical Committee of China Agricultural University. All institutional and national guidelines for the care and use of animals were followed.
Conflict of interest
Animal and Human Rights Statement
-

Table Supplementary Table S1. Primers used in this study

Table Supplementary Table S2. Molecular weight (MW) information for PRRSV nsps involved in this study

















 DownLoad:
DownLoad: