HTML
-
The human interferon-inducible myxovirus resistance proteins (MxA and MxB) belong to the dynamin superfamily of large GTPases. They share 63% amino acid sequence identity and have identical structure and domain architecture, including the N-terminal GTPase (G) domain, the bundle signaling element (BSE) and the stalk domain (Haller et al. 2015). Human MxA has long been established as a potent interferon-induced restriction factor for a wide variety of viruses, though it does not inhibit lentiviruses such as HIV-1. The anti-viral activity of MxA depends on both oligomerization and GTPase activity, including GTP binding and GTP hydrolysis (Verhelst et al. 2013). It has been proposed that oligomerization of MxA on viral substrates activates the GTPase and that GTP hydrolysis induces a conformational change in the oligomer (Dick et al. 2015; Nigg and Pavlovic 2015). Recent study indicated that functions of MxA rely on domain rearrangements coupled with GTP hydrolysis cycles. GTP binding is associated with the lever-like movement of structures adjacent to the GTPase domain, while GTP hydrolysis returns MxA to its resting state (Chen et al. 2017).
MxB has now established broad-spectrum antiviral profile effective against HIV-1 (Goujon et al. 2013; Kane et al. 2013; Liu et al. 2013) and herpesviruses (Crameri et al. 2018; Schilling et al. 2018), as well as HTNV (Li et al. 2019), HCV (Yi et al. 2019) and HBV (Wang et al. 2020). Like MxA, MxB oligomerization is also important for its anti-viral functions. MxB has several assembly interfaces that are critical for HIV-1 restriction. Stalk domains of MxB provide the appropriate spacing for efficient interaction with hexamer interfaces in the HIV-1 capsid lattice (Fribourgh et al. 2014; Goujon et al. 2015; Kong et al. 2016; Alvarez et al. 2017). GTP binding and hydrolysis can coordinately signal through the BSE to the stalk, resulting in conformational changes. In this regard, the Mx GTPase resembles a Swiss Army knife, exposing different antiviral interfaces depending on varying conformations (Haller et al. 2015). GTPase activity is critical for MxB to exert its cellular functions (King et al. 2004). However, the role of MxB GTPase activity in viral inhibition is not well understood. It is confusing that the GTPase activity is required for its anti-viral effects against herpesviruses and HBV but not HIV-1 (Goujon et al. 2013; Kane et al. 2013; Matreyek et al. 2014; Crameri et al. 2018; Meier et al. 2018; Schilling et al. 2018; Wang et al. 2020).
In this study, to characterize the role of GTP binding and GTP hydrolysis in the MxB-mediated inhibition of HIV-1, we therefore genetically separated these two functions and evaluated their contributions to restriction. Our results provide novel insights to understand the mechanism of the anti-HIV-1 action of MxB based on GTP binding and GTP hydrolysis.
-
Human MxB (NM_002463.1) was cloned into the pCMV-C-HA vector (Clontech, USA) using BamHI and XhoI site to generate the HA-labeled MxB expression plasmid. Plasmids expressing the MxB mutants were created by site directed mutagenesis. Constructs were confirmed by sequencing analysis.
The HIV-1 NL4-3 molecular clone (pNL4-3) and HIVLuc construct (pNL4-3-Luc-R-E-) were obtained from the NIH AIDS reagent program.
-
Human 293T cells and human HeLa cells were grown on DMEM supplemented with 10% fetal bovine serum (FBS) (Gibco, USA) and 1% (w/v) penicillin/streptomycin.
-
Vesicular stomatitis virus G protein (VSV-G)-pseudotyped firefly luciferase protein (Luc)-encoding HIV-1 stocks were prepared in 293T cells by co-transfection with pNL4-3- Luc-R-E- and pMD2.G using a 3:1 ratio. Virus containing supernatant were collected 48 h post transfection, clarified by low-speed centrifugation (300 ×g, 5 min), and filtered through 0.45 μm pore size sterile filters. Viral particles were then normalized by HIV-1 p24 ELISA using p24 ELISA Kit (Zepto Metrix, USA). For infection assays, 1 × 105 HeLa cells were transduced in 24-well plate in triplicate and were inoculated with virus at 37 ℃ for 4 h in the presence of 8 μg/mL of polybrene, after which virus containing medium was removed and replaced with new medium. Infectivity was measured 48 h post infection and luciferase activity was measured using the Steady-Glo kit (Promega, Germany).
-
HeLa cell cultures were inoculated with HIV-1 particles pseudotyped with VSV-G as indicated. Twenty-four hours post infection, cells were detached with trypsin, pelleted and washed once with 1× PBS. The cells were then lysed with TRIZOL (Invitrogen, USA). Total DNA was isolated according to the manufactures protocol and 100 ng of each sample was used for real-time PCR analysis using 2× Taq-Man Fast qPCR Master Mix (BBI Life Sciences, China). The second strand transfer products (Late RT) were detected using primers U5 forward, 5'-CAGACCCTTTTAGT CAGTGTGGAA-3 and U5 reverse, 5'-CTCTGGCTTTACT TTCGCTTTCA-3', with U5 probe, 5'-(FAM)-TCTCTAGC AGTGGCGCCCGAACA-(TAMRA)-3'. 2-LTR circle products were detected using primers 2-LTR forward, 5'-AACTAGGGAACCCACTGCTTAAG-3' and 2-LTR reverse, 5'-TCCACAGATCAAGGATATCTTGTC-3', with 2-LTR probe 5'-(FAM)-ACACTACTTGAAGCACT CAAGGCAAGCTTT-(TAMRA)-3' (Kitagawa et al. 2008). Integrated (provirus) DNA was analyzed using Alu PCR (Butler et al. 2001). In brief, 16 cycles of pre-amplification (15 s at 94 ℃, 15 s at 55 ℃, 100 s at 72 ℃) were conducted with Taq DNA Polymerase (Invitrogen, USA) using 600 nmol/L of U5 forward primer and 100 nmol/L of genomic Alu reverse primer, 5'-TGCTGGGATTACAGGCGTGAG-3'. Second-round qPCR was performed on preamplification products using the U5 forward and U5 reverse primers with the U5 probe. qPCR reactions were performed in triplicate, in universal PCR master mix using 900 nmol/L of each primer and 250 nmol/L probe. After 10 min at 95 ℃, reactions were cycled through 15 s at 95 ℃ followed by 1 min at 60 ℃ for 40 repeats. Quantitative RT-PCR was performed using Bio-Rad CFX96 instrument (BioRad, USA). GAPDH was also quantified in each sample to normalize HIV-1 cycle threshold (Ct) values to generate ΔΔCt values.
-
For co-immunoprecipitation analysis, 293T cells were washed with ice-cold PBS, lysed in hypotonic lysis buffer (10 mmol/L Tris–HCl pH 8.0, 10 mmol/L KCl, 1 mmol/L EDTA, 1× protease inhibitor cocktail) by homogenizer. Lysate was centrifuged at 1500 ×g for 5 min at 4 ℃ and the supernatant was immunoprecipitated with rabbit antiNUP358 antibody (Abcam, ab64276, USA), rabbit antiNUP98 (CST, C39A3, USA), rabbit anti-HA (CST, C29F4, USA) or control rabbit IgG (ABclonal, AC005, China), by incubating with Protein-G Sepharose (GE Healthcare, USA) at 4 ℃ with gentle agitation for 4 h. The immunoprecipitates were then washed with PBS twice, followed by a final wash with lysis buffer before mixing with 2× SDS-PAGE loading dye.
-
Cell lysates were prepared by lysing cells with Radio Immunoprecipitation Assay (RIPA) lysis buffer containing protease inhibitor cocktail (Roche, China) for 15 min on ice. Following incubation, lysates were spun down at 12, 000 ×g for 5 min and supernatant was collected for western blot analysis. In brief, 5× SDS loading buffer were added to the lysed sample and incubated at 100 ℃ for 5 min. Protein concentration was measured using Pierce BCA protein assay kit (Thermo Scientific, USA) and equal amount of protein was loaded into an 8% polyacrylamide gel for SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Upon separation, the proteins were transferred to PVDF membrane (Merckmillipore, Germany) using TransBlot Turbo transfer system (Bio-Rad, USA). The primary antibodies used were mouse anti-HA (BioLegend, #901513, 1:2000), mouse anti-p24 (Abcam, ab9071, 1:2000), mouse anti-GAPDH (Cwbiotech, CW0100M, 1:5000), rabbit anti-NUP358 (Abcam, ab64276, 1:2000) and rat anti-NUP98 (Abcam, ab50610, 1:2000). Detection were performed using the Horseradish Peroxidase (HRP) conjungated-Goat anti-mouse or anti-rabbit IgG secondary antibodies (Cwbiotech, China) at dilution of 1:5000, followed by chemiluminescence detection using Pierce ECL Plus Western blotting substrate (Thermo Scientific, USA) in the ChemiDoc Imaging System (Tanon, China).
-
Cells cultured on 20-mm-diameter Glass Bottom Cell Culture Dish were fixed with 4% paraformaldehyde for 20 min. After washed three times with PBS, the cells were permeabilized with 0.25% Triton X-100 for 10 min. The permeabilized cells were blocked with PBS containing 3% BSA for 1 h at 37 ℃ and stained with indicated primary antibodies diluted in blocking buffer for overnight at 4 ℃. Then, cells were washed three times with PBS, and incubated with fluorescently conjugated secondary antibody diluted in blocking buffer for 1 h at 37 ℃ and again washed with PBS three times. Cells were stained with Hoescht 33342 (Invitrogen, USA) diluted to a concentration of 1 mg/mL for 3–5 min, following three washes with PBS. Subsequently, samples were mounted for fluorescent microscopy by using the Antifade Mounting Medium (Beyotime, China). Images were obtained with Leica confocal microscope (Leica-LCS-SP8-STED, Germany) using a 63× objective and were analyzed with Leica Application Suite X software.
The following antibodies and dilutions were used for immunofluorescence studies: rabbit anti-NUP358 (Abcam, ab64276, 1:2000), rabbit anti-NUP98 (CST, C39A3, 1:100), mouse anti-HA (BioLegend, #901513, 1:1000), rabbit anti-HA (CST, C29F4, 1:800) and mouse anti-p24 (Abcam, ab9071, 1:200) antibodies. The following fluorescently labeled secondary antibodies were used: Dylight 594-conjugated goat anti-mouse IgG antibody (Abbkine, A23440, 1:500) and Dylight 488-conjugated goat antirabbit IgG (Abbkine, A23210, 1:500).
-
Where appropriate, data were expressed as mean ± SD of triplicate cultures. Individual statistical tests were specified within the figure legends. Count data with more than two groups were analyzed by Kruskal–Wallis test. All other grouped data were analyzed by one-way ANOVA. Statistical analysis was performed with GraphPad InStat statistical software (GraphPad Software, USA). Statistical significance was defined as P < 0.05.
Plasmids and Viral Vectors
Cell Lines
Virus and Infection
Real-Time PCR to Detect HIV-1 Reverse Transcription Products
Co-Immunoprecipitation Assays (Co-IP)
Western Blotting
Immunofluorescence Confocal Microscopy
Statistical Analysis
-
Previous work has proposed a model whereby multiple components of the nuclear import machinery and nuclear pore complex (NPC) help position MxB at the nuclear envelope to promote MxB-mediated restriction of HIV-1 (Dicks et al. 2018). In order to investigate the role of GTPase activity in MxB restriction to HIV-1 replication, we introduced mutations into the GTPase domain (Fig. 1A) and subsequently tested the mutant MxB proteins for their association with NUPs in HeLa cells (Fig. 1B). Indirect immunofluorescence analysis of transiently transfected HeLa cells revealed that wild-type MxB mainly localized along the nuclear rim and also in a punctate cytoplasmic pool, exhibited significantly colocalization with NUP358 and NUP98. In contrast to wild-type MxB, the mutant MxB proteins exhibited a different pattern of localization. GTP-binding defect (K131A) or deletion of the nuclear localization signal (NLS) resulted in cytoplasmic localization and reduced colocalization with NUPs. MxBT151A, which loses the enzymatic activity of GTP hydrolysis but retains GTP binding ability, accumulated almost exclusively to the nuclear envelope. It displayed similar patterns of colocalization with NUP358 as wild-type MxB, but had reduced colocalization with NUP98 (Fig. 1C). In vitro Co-IP assays confirmed the importance of GTP binding in MxB-NUP358 interaction, whereas intact GTPase activity in MxB-NUP98 interaction (Fig. 1D).
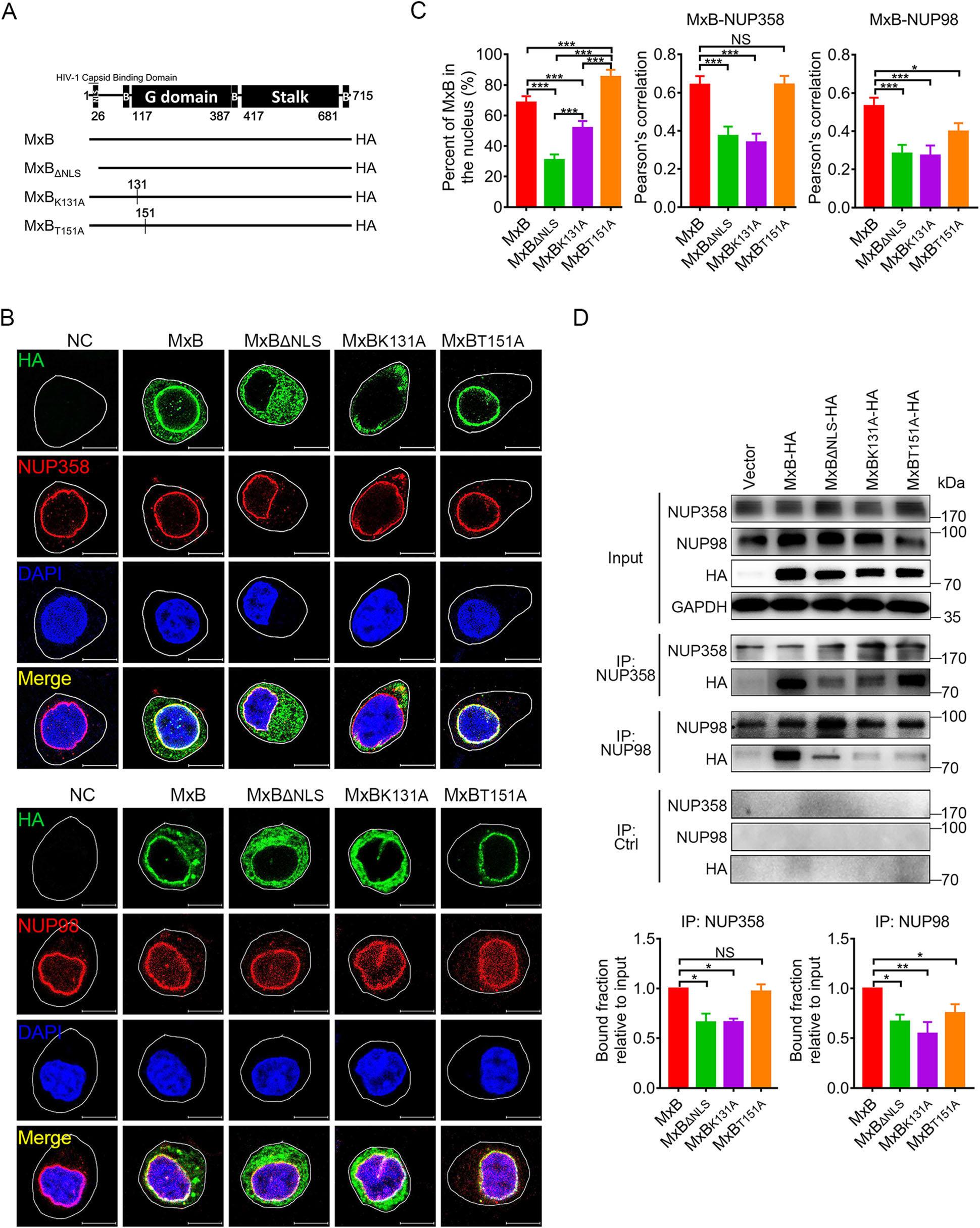
Figure 1. GTPase activity affects MxB nuclear location and its association with NUPs. A Wild-type human MxB protein was depicted in the top figure. The numbers of amino acid residues at the boundaries of the MxB domains were indicated. Different MxB variants were shown. B Immunofluorescence stained NUP358 or NUP98 (red). HA-tagged indicated MxB proteins (green) and DAPI-stained DNA (blue) in HeLa cells. Scale bar, 10 μm. C Distribution of MxB proteins in the nucleus and Pearson correlation coefficient values for colocalization of indicated MxB proteins and NUP358, NUP98 were analyzed by image analysis software. Individual measurements with mean ± SD were shown. Data were representative of two independent experiments. For each comparison, 25 or more cells were randomly selected, Kruskal–Wallis test was performed. D Co-immunoprecipitation analysis of interaction between MxB and NUP358/NUP98. The quantification of bound fractions relative to the input was calculated for each experiment. Bars indicate mean ± SD, n = 3 independent experiments, one-way ANOVA was performed (NS, not significant, *P < 0.05, **P < 0.01 and ***P < 0.001).
These observations indicated that in addition to NLS, GTPase activity of MxB also contributes to its nuclear location and interaction with NUPs. GTP binding facilitates MxB interaction with the cytoplasmic face of nuclear pore (NUP358), while GTP hydrolysis does not. Both GTP binding and GTP hydrolysis facilitate MxB interaction with the inner side of nuclear pore (NUP98). MxB probably releases from NUP358 then moves to NUP98, and this process is strictly associated with GTP hydrolysis of MxB.
-
To characterize the role of GTPase activity in the MxB-mediated inhibition of HIV-1, we next tested the mutant MxB proteins for their potential abilities to inhibit pseudotyped HIV-1 replication in HeLa cells. Consistent with previous reports (Goujon et al. 2013; Kane et al. 2013; Matreyek et al. 2014; Meier et al. 2018), MxB potently reduced pseudotyped HIV-1 infection while the short isoform (MxBΔNLS) had no observable anti-HIV-1 activity. Mutations K131A and T151A exhibited modest anti-HIV-1 activity (Fig. 2A). We also measured viral reverse transcripts representing three phases of HIV-1 replication: the second strand transfer products (Late RT), 2-long terminal repeat (LTR) circular DNA (a marker for viral cDNA nuclear localization) and integrated (provirus) DNA respectively. As previously shown (Goujon et al. 2013), MxB reduced the levels of viral 2-LTR circles and integrated DNA but not Late RT DNA. Conversely, MxBΔNLS did not measurably affect the accumulation of all HIV-1 DNAs. Both defects in GTP binding (K131A) and GTP hydrolysis (T151A) showed a significant decrease of 2-LTR circles and integrated DNA, but less strong compared to MxB (Fig. 2B). These indicated that GTP binding and hydrolysis may contribute to inhibiting the nuclear import of viral replication complexes. However, the most intriguing thing was that only MxBK131A reduced the Late RT DNA (Fig. 2B). The accumulation of MxBK131A in the cytoplasm may provide the opportunity to interfere with reverse transcription. Different exhibitions of MxB mutations on three viral DNA analysis indicated distinct restriction mechanism based on GTP binding and GTP hydrolysis.
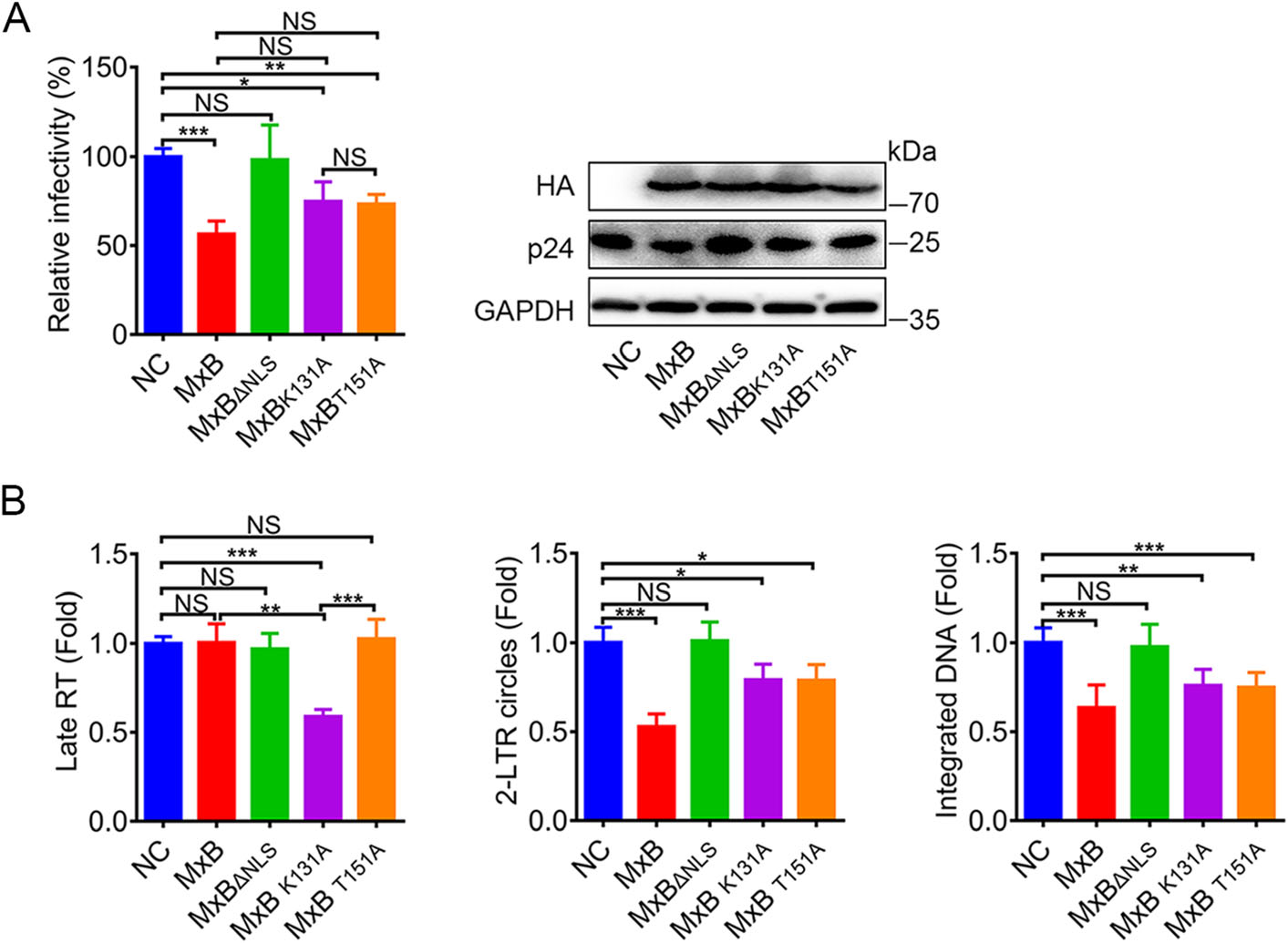
Figure 2. MxB restricts HIV-1 replication via distinct mechanism based on GTP binding and GTP hydrolysis. A Infectivity of VSV-G pseudotyped HIV-1 luciferase reporter virus in HeLa cells transfected with plasmid expressing wild-type MxB-HA or the indicated mutant MxB-HA protein. B qPCR analysis of the late viral DNA, 2-LTR circle DNA and integrated proviral DNA in HeLa cells transfected with plasmid expressing wild-type MxB-HA or the indicated mutant MxB-HA protein. Bars indicate mean ± SD, n = 3 independent experiments, one-way ANOVA was performed (NS, not significant, *P < 0.05, **P < 0.01 and ***P < 0.001).
-
MxB inhibits HIV-1 replication by interacting with the HIV-1 capsid (CA) via its N-terminal and GTPase (G) domains (Betancor et al. 2019). To illustrate the role of MxB GTPase activity in viral target recognition, we investigated the association of MxB with HIV-1 cores by immunofluorescence confocal assays. The percentage of p24 that colocalized with MxB was calculated. As shown in Fig. 3A, deletion of NLS domain or mutation in GTP binding region (K131A) resulted less colocalization of MxB and HIV-1 CA. Similarly, in Co-IP analysis, MxBΔNLS and MxBK131A showed reduced MxB-CA interaction (Fig. 3B), suggesting that GTP binding facilitates MxB-CA interaction, while GTP hydrolysis does not. These results highlighted the significant role played by both NLS domain and GTP binding activity in maximizing efficient viral capsid recognition.

Figure 3. MxB NLS and GTP binding region are important to the interaction between MxB and HIV-1 cores. A Immunofluorescence stained HA-tagged indicated MxB proteins (red), HIV-1 capsid protein p24 (green) and DAPI (blue) in HeLa cells at 6 h post VSV-G pseudotyped HIV-1 luciferase reporter virus infection. The arrows indicated the p24 associated with MxB. Scale bar, 10 μm. The percentage of p24 associated with MxB was quantified by image analysis software in the right. Individual measurements with mean ± SD were shown. Data were representative of two independent experiments. For each comparison, 25 or more cells were randomly selected, Kruskal–Wallis test was performed. B Coimmunoprecipitation analysis of interaction between MxB and HIV-1 CA. The quantification of bound fractions relative to the input was calculated for each experiment in the right. Bars indicate mean ± SD, n = 3 independent experiments, oneway ANOVA was performed (NS, not significant, *P < 0.05, **P < 0.01 and ***P < 0.001).
-
Both NUP358 and NUP98 have been found to bind HIV-1 CA and are involved in HIV-1 pre-integration complex (PIC) nuclear import (Ebina et al. 2004; Zhang et al. 2010; Di Nunzio et al. 2013). Since MxB could bind with NUP358 and NUP98 as well as HIV-1 CA (Fig. 1, Fig. 3), we therefore wondered whether MxB affects the NUP-sbased HIV-1 nuclear import. We expressed MxB proteins in HeLa cells (Fig. 4A), and assessed the degree of colocalization between HIV-1 CA and NUPs post pseudotyped HIV-1 infection by immuno-fluorescence confocal assays (Fig. 4B). As shown in Fig. 4C, wild-type MxB reduced nuclear CA accumulation and consistently with less CA-NUP358/98 colocalization. Both MxBΔNLS and MxBK131A failed to prevent CA-NUP358/98 colocalization and nuclear CA accumulation, which most likely due to their less interaction with NUPs and HIV-1 CA (Fig. 1, Fig. 3). It was consistent that T151A mutation led to accumulation of both HIV-1 CA and MxB per se on NUP358 but less to NUP98, suggesting MxBT151A arrested both CA and itself on NUP358, thus disrupting HIV-1 nuclear import. These differences between wild-type MxB and GTPase-defective mutants indicated MxB inhibited PIC nuclear import at different transport stage based on GTPase activity (Fig. 5).
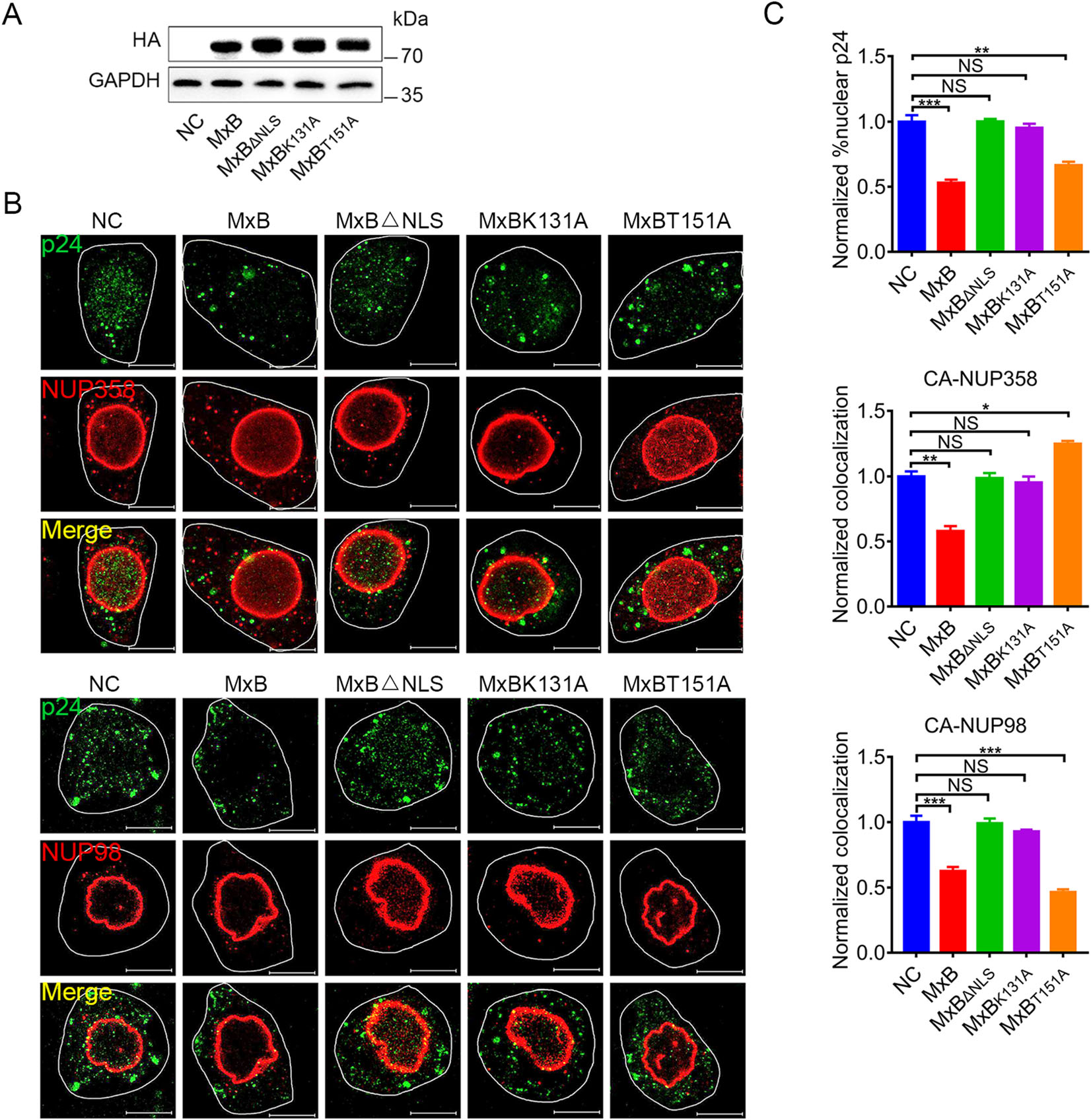
Figure 4. MxB affects the association between NUPs and HIV-1 cores dependently on GTPase activity. A Expression of wild-type MxB-HA or the indicated mutant MxB-HA protein in HeLa cells were monitored by western blot. B Immunofluorescence stained NUP358 or NUP98 (red) and HIV-1 capsid protein p24 (green) in HeLa cells at 6 h post VSV-G pseudotyped HIV-1 luciferase reporter virus infection. Scale bar, 10 μm. C Quantification of the percentage of the CA detected in the nucleus and the percent CA colocalizing with NUP358, NUP98. Individual measurements with mean ± SD were shown. Data were representative of two independent experiments. For each comparison, 25 or more cells were randomly selected, Kruskal– Wallis test was performed (NS, not significant, *P < 0.05, **P < 0.01 and ***P < 0.001).
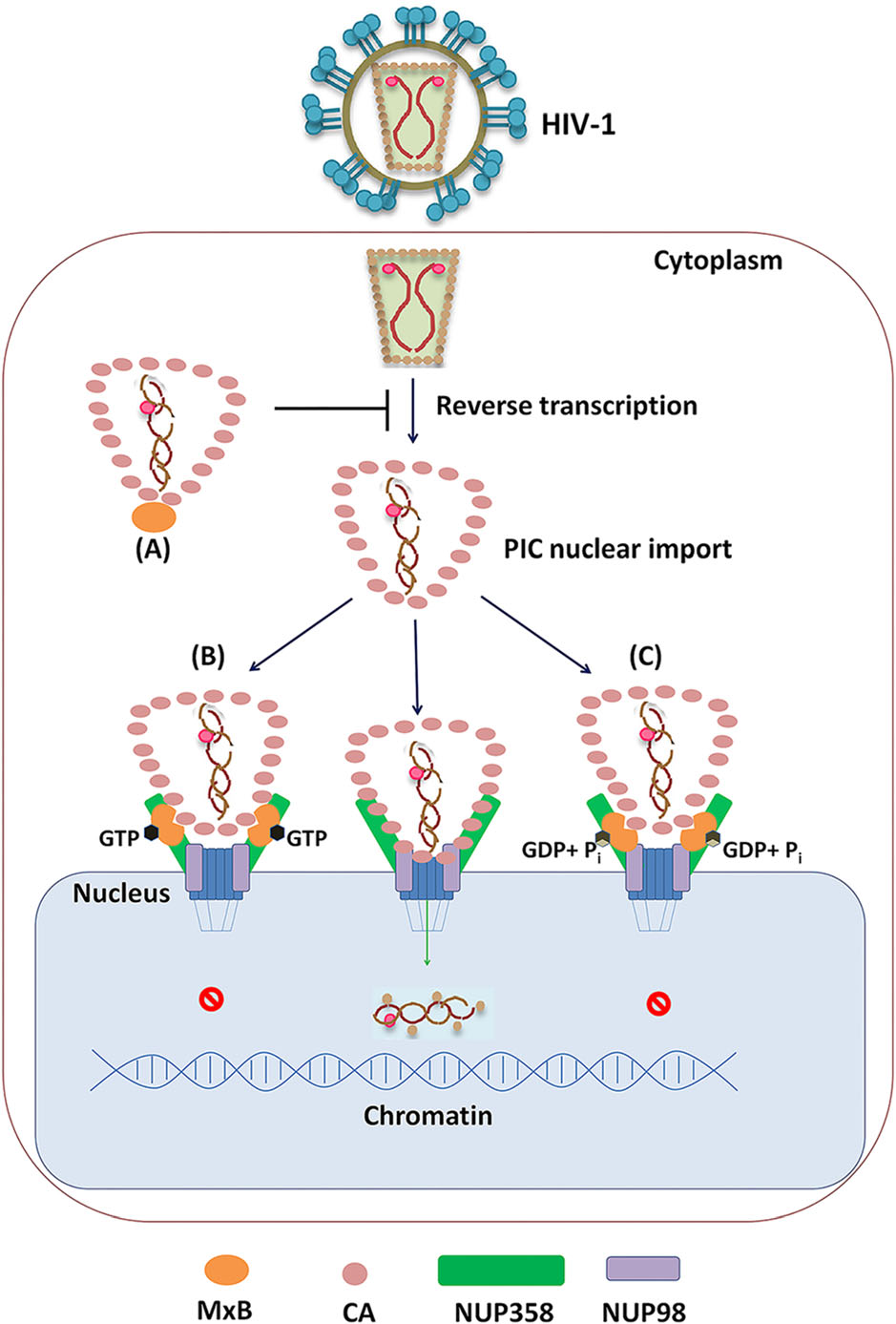
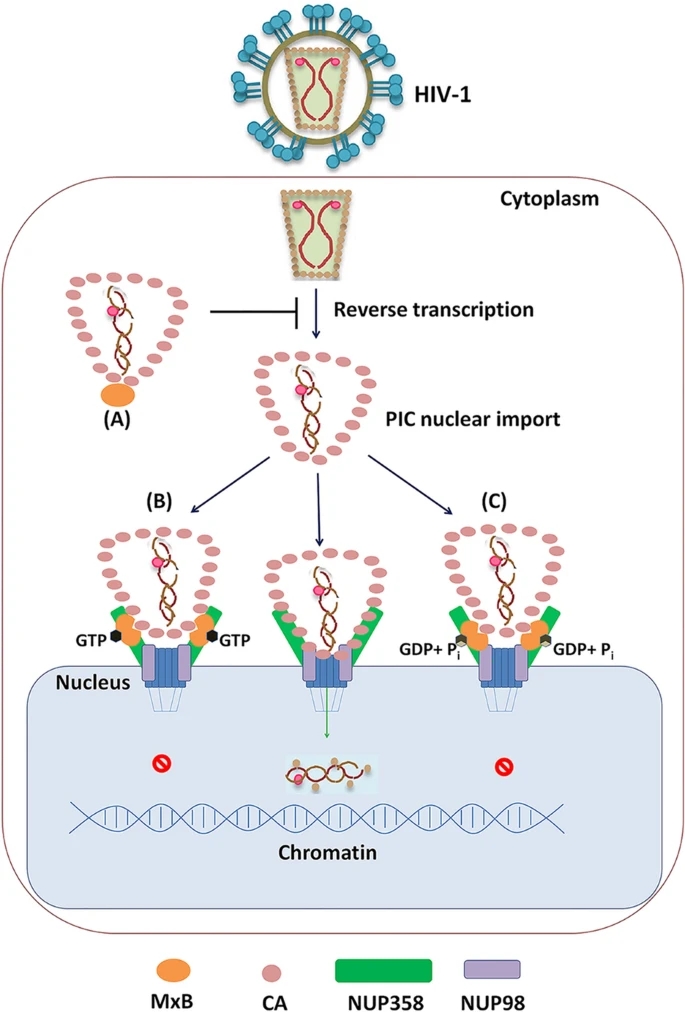
Figure 5. Distinct mechanism for MxB to inhibit HIV-1 replication based on GTP binding and GTP hydrolysis. A When MxB does not bind GTP (MxBK131A), it mainly locates at cell cytoplasm, inhibits HIV-1 reverse transcription process, resulting decreased HIV-1 replication. B GTP-bound MxB (MxBT151A) mainly accumulates on NUP358 but less on NUP98, it can recognize HIV-1 CA and lock HIV-1 CA to NUP358, inhibiting PIC nuclear import. C Wild-type MxB can bind GTP, then hydrolyzes GTP to GDP and Pi. MxB accumulates on both NUP358 and NUP98, reduces the CA-NUP358/ 98 interaction, inhibiting PIC nuclear import.
GTPase Activity Affects Mxb Nuclear Location and Its Association with NUPs
MxB Restricts HIV-1 Replication Via Distinct Mechanism Based on GTP Binding and GTP Hydrolysis
MxB NLS and GTP Binding Region are Important to the Interaction Between MxB and HIV-1 Cores
MxB Affects Association Between NUPs and HIV-1 Cores Dependently on GTPase Activity
-
Human MxB was reported to target the HIV-1 capsid (CA) after cell entry (Busnadiego et al. 2014; Fribourgh et al. 2014; Kong et al. 2014; Buffone et al. 2015), to prevent uncoating (Fricke et al. 2014), nuclear import of the viral pre-integration complex (PIC) and subsequent chromosomal integration of the proviral DNA into the host genome, but did not affect reverse transcription (Goujon et al. 2013; Kane et al. 2013; Liu et al. 2013; Matreyek et al. 2014; Haller et al. 2015). Recent study emphasized the importance of the GTPase (G) domain of MxB in the inhibition of HIV-1 replication (Betancor et al. 2019). Similarly, it has been described that the GTP binding and hydrolysis activity of MxB are necessary for the inhibition of herpesviruses (Crameri et al. 2018; Schilling et al. 2018), while GTP binding but not GTP hydrolysis activity is required for anti-HBV activity (Wang et al. 2020). However, mutant MxB proteins deficient for GTPase activity maintain anti-HIV-1 function (Goujon et al. 2013; Kane et al. 2013; Matreyek et al. 2014; Meier et al. 2018). The role of GTPase activity of MxB in the inhibition of HIV-1 is still less clear. In this work, we revisited the role of GTPase activity of MxB for the control of HIV-1 replication. We showed that, in addition to NLS, GTPase activity of MxB also affected its function at unique steps in HIV-1 life cycle. Both MxB and MxBT151A blocked the nuclear accumulation and integration of HIV-1 reverse transcripts but not reverse transcription. However, MxBK131A, deficient in GTP binding, could interfere with HIV-1 reverse transcription. These findings strongly suggested that GTP binding to MxB facilitated HIV-1 reverse transcription, but GTP hydrolysis did not.
MxB was first identified to function at the gate of nuclear pore for cytoplasmic-nuclear transport (King et al. 2004). Recently, it has been reported that MxB interacts with multiple components of the nuclear pore complex, including NUP98 and NUP214 (Dicks et al. 2018). Herein, we confirmed associations between MxB and NUP358/98, and further found that GTP binding facilitated NUP358- MxB interaction, while GTP hydrolysis reduced that interaction. Importantly, MxB need intact GTPase activity for NUP98-MxB interaction. In light of the location of MxB and NUPs, we propose a model in which GTP binding helps recruit MxB to NUP358, then MxB can be subsequently translocated to NUP98 together with GTP hydrolysis. MxB could target HIV-1 cores through its NLS and G domain during PIC nuclear import, which also required its dimerization. A recent study indicated GTPase activity may be involved in the organization of MxB oligomerization (Alvarez et al. 2017). These findings indicated that the GTPase activity might affect MxB-CA association by regulating the dimerization of MxB.
Previous studies surmised that MxB inhibits the nuclear import of HIV-1 PIC and the integration of proviral DNA by influencing viral capsid interaction with NUPs (Staeheli and Haller 2018). Probably due to distinct MxB-NUPs and MxB-CA interactions, GTPase activity deficient MxB mutants exhibited anti-HIV-1 function through different mechanisms. Actually, GTP-binding defect (K131A) reduced MxB-NUP358/98 interactions as well as MxB-CA interactions, presented less MxB on NUP358/98, which reduced impacts on CA-NUP358/98 interactions. We suppose that the GTP-binding deficient MxB inhibit HIV-1 reverse transcription process, which contributes predominantly to the decreased HIV-1 replication (Fig. 5A). The second, GTP-hydrolysis defect (T151A) led to MxB accumulate on NUP358 but less on NUP98, locked HIV-1 CA to NUP358, thus inhibiting PIC nuclear import (Fig. 5B). Finally, in the case of wild-type MxB, intact GTPase activity ensured MxB interact with NUPs and CA properly, as a result, the wild-type MxB significantly reduced the CA-NUP358/98 interaction, which led to interfered HIV-1 replication (Fig. 5C).
In conclusion, we provide details of interactions among MxB, HIV-1 CA and NUPs at NPC, and how MxB acts differently at this site based on its GTPase activity. We could explain how MxB inhibits HIV-1 ''independently'' on its GTPase activity, just because of the unique mechanism utilized.
-
We are grateful to the Medical Research Center for Structural Biology, Wuhan University School of Basic Medical Sciences for providing experimental facilitates. This work was supported by the National Science Foundation of China (81271818 and 81471940 to YF, and 81471941, 81871659 and 81828005 to WH).
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
-
LX carried out all the experiments, collected data and wrote this manuscript. ZJ, CZ, YW and YZ helped to finish the experiments. LX, WH, and YF designed the experiments, analyzed the data, checked and finalized the manuscript. All authors read and approved the final manuscript.















 DownLoad:
DownLoad: