HTML
-
In December 2019, SARS-CoV-2, a novel coronavirus (Wu et al. 2020; Zhou P et al. 2020), caused an outbreak of acute pneumonia in Wuhan, China (Lu et al. 2020; Singhal 2020; Yang S et al. 2020; Ye et al. 2020; Zhou F et al. 2020). The World Health Organization (WHO) declared SARS-CoV-2 infection outbreak as a "public health emergency of international concern" on January 30, 2020. Later on March 11, WHO defined COVID-19 as a pandemic (WHO 2020a). Until June 11, it has been reported globally 7, 273, 958 COVID-19 cases and 413, 372 deaths in 215 countries (WHO 2020b). The complete clinical picture of COVID-19 is only partially understood. COVID-19 cases have ranged from very mild (including asymptomatic), mild, moderate to severe and critical severe including illness resulting in death. COVID-19 common symptoms include fever, cough and shortness of breath. Muscle pain, sputum production and sore throat are less common. While the majority of the cases show mild symptoms, some progress to severe pneumonia and multi-organ failure including lung, heart, and kidney. The rate of deaths per number of diagnosed cases is estimated to be 3.4% but varies by age and other health conditions (Lu et al. 2020; Singhal 2020; Wang T et al. 2020; Wu and McGoogan 2020; Yang S et al. 2020, Yang X et al. 2020; Ye et al. 2020; Zhang C et al. 2020, Zhang H et al. 2020; Zhou C et al. 2020, Zhou F et al. 2020; Zou L et al. 2020). The mechanisms of how SARS-CoV-2 cause severe multi-organ failure are largely unknown.
Acute kidney injury (AKI) is the second leading frequent organ damage in coronavirus infected cases, which goes just after respiratory system injuries (Chafekar and Fielding 2018). The epidemiological and clinical studies showed that 3%–19% COVID-19 cases were reported with AKI (Huang et al. 2020; Li et al. 2020). The latest study also reported the detection of viral RNA in urine samples of COVID-19 patients (Guan et al. 2020). However, whether AKI of COVID-19 patients are caused by coronavirus-induced cytopathic effect or immunopathogenic damage remains unclear.
Previous studies have shown that SARS-CoV-2 enters cell through its receptor, angiotensin converting enzyme 2 (ACE2) (Zhou P et al. 2020). Single-cell RNA sequencing data analysis showed that ACE2 is expressed in kidney proximal convoluted tubules (Li et al. 2020; Pan et al. 2020). These data indicated that renal tubule cells could be the potential host cells targeted by SARS-CoV-2. Due to the lack of specific detection of ACE2 mRNA and protein expression in human kidney tubule cells, it is hard to confirm the direct infection of SARS-CoV-2. Since expression of ACE2 may play roles in the regulation of renal function, it's of interest to uncover the mechanisms by which SARS-CoV-2 infection damages kidney function.
Animal models for study of SARS-CoVs include African green monkeys, macaques and mice (Kong et al. 2009; Smits et al. 2011). However, there are some limitations because of species difference. The viral tropism correlates to expression of the viral receptor of host cells and intracellular restriction as well. For example, human influenza viruses preferentially bind to α2-6-Gal linked sialic acids, while avian influenza viruses use α2-3-Gal linked sialic acids as receptors (Guo et al. 2009; Chan et al. 2013).
Human kidney proximal tubule epithelial cells (KPTECs) are important for many functions of human kidney, such as clearing blood and resorption of essential end-products from metabolism, including protein, sodium, etc. (Smith et al. 2006). These cells are highly useful for studying basic cell biology and modeling kidney diseases (Phillips 2003). However, primary KPTECs in vitro undergo a very limited number of population doublings (PDs), thus it would be difficult to obtain reproducible results due to differences of primary cells. Previous studies have been focused on the immortalization of KPTECs using viral oncogenes HPV16 E6/E7, or a hybrid adeno-12-SV40 virus, or SV40 and hTERT (Ryan et al. 1994; Racusen et al. 1997; Kowolik et al. 2004; Orosz et al. 2004; Wieser et al. 2008). However, due to genetic manipulation, these immortalized cells and traditional cancer cell lines largely loss differentiation capacity and normal physiological function. There is an unmet need for normal epithelial cells for disease modeling.
Recently, conditional reprogramming (CR) allows for indefinite proliferation of human epithelial cells with no transduced viral or cellular genes (Liu et al. 2012, 2017). CR cells (CRCs) maintain differentiation potential and lineage function in vitro (Liu et al. 2012; Suprynowicz et al. 2012; Zhu et al. 2017). In this study, we firstly established long term cell cultures of KPTECs using 2D CR and 3D organoids technologies, which maintained the lineage function, and the ability to differentiate and repair DNA damage. They also did not have transforming property as cancer cells did. Importantly, these cells expressed endogenous specific transporters SLC34A3 and cubilin, and ACE2. The latter is a receptor for entry of SARS-CoV and SARS-CoV-2. In contrast, cancer cell line did not express these proteins endogenously. Very interestingly, ACE2 expression was around twofold higher in 3D organoids compared to that in 2D CR condition. This integrated 2D CR and 3D organoid cultures provide a novel ex vivo model to study kidney functions, innate immune response of kidney cells to viruses, and a novel platform for drug discovery and safety evaluation.
-
Cryopreserved primary KPTECs were purchased from Lonza (Catalog #: CC-2553). Cells were cultured in CR condition on irradiated 3T3-J2 fibroblasts as described previously (Liu et al. 2012, 2017) and passaged in Dulbecco's modified Eagle medium (DMEM)/F12 containing 5 μmol/L Y-27632 (YongTech, Shenzhen). CR KPTECs were trypsinized in two steps by using 0.05% Trypsin–EDTA. The initial 30 s trypsinization to remove feeders was followed by a wash of PBS and another 3 min trypsinization to detached cells. Cells were subsequently reseeded at a 1:4–6 ratio in the CR condition and cultured for 3–4 days before passaging.
-
KPTEC-CRC cultures were established in Matrigel Organoids 3D condition as previously described (Liu et al. 2012; Saenz et al. 2014). Briefly, 40 µL of cold Matrigel (BD Bioscience Cat #354230) was added to the surface of sterile pre-chilled, slide chambers (Lab-Tek Ⅱ Chamber Slide, ThermoFisher Scientific Inc.). The slides were then incubated at 37 ℃ for 15 min to allow polymerization of the substratum. Subsequently, 3 × 104 cells/mL were added to the slides and resuspended in assay media (YongTech, Shenzhen) containing 2% FBS, 2% Matrigel, and 20 ng/mL EGF and incubated at 37 ℃ until 3D organoid formation. 0.5 μg/mL DAPI (D3571) was used to stain the nuclei. Bright field microscopy, confocal microscopy, immunostaining were carried out as described previously (Liu et al. 2012; Saenz et al. 2014).
-
Cells were grown on sterile glass cover slips and treated with 5 Gy of gamma irradiation and fixed in 4% (w/v) paraformaldehyde at indicated time points, permeabilized with 0.1% saponin in PBS containing 0.2% gelatin, and labeled with the primary (mouse anti-p63, Santa Cruz, sc-863; Goat polyclonal ACE2 antibody, R & D systems, Abingdon, UK) and secondary (Alexa Fluor 488 donkey anti-mouse IgG, anti-goat Alexa Fluor 488, Molecular Probe) antibodies, according to the manufacturer's protocol. 0.5 μg/mL DAPI (D3571) was used to stain the nuclei DNA. A Zeiss Axioskop microscope (Carl Zeiss, Inc., Thornwood, NY) equipped with a × 63 objective lens and a Hammamutsu charge-coupled device camera was used to image the cells. Images were processed using Openlab 3.0.7 software.
-
Anchorage-independent growth assays with 4 × 104 cells in 0.3% low-melting point agarose were performed according to the previous study (Feng et al. 2015). Colony images were captured and analyzed with the EVOS flat screen microscope (Life Technologies).
-
Total cellular RNAs were extracted using TRIzol Reagent (ThermoFisher) and real time RT-PCR was performed as described previously (Wang et al. 2018). The primer sequences for detection of ACE2 mRNA were 5′-CATTGGAGCAAGTGTTGGATCTT-′3 and 5′-GAGCTAATG-CATGCCATTCTCA-3′; for SLC34A3 were 5′-CGCAGGCGCCCGACATCC-3′ and 5′-ACCTGTTTGCGGGCACGGAGCTCA-3′; for human cubilin were 5′-GCTCATCCAGGCTCCCGACTCTAC-3′ and 5′-TTGAAGCCTGCCCTGGTTACACTG-3′ (Wieser et al. 2008). β-Actin was used as an internal control (5'-GAGCACAGAGCCTCGCCTTT-3', 5'-TCATCATCCATGGTGAGCTGG).
-
A total of 5 × 103 HeLa cells in complete DMEM or CR PKTECs were seeded into 96-well plate. HEK293T cells were co-transfected with pSpike (SARS-CoV S), psPAX, and lenti-Luc for 4 h and pseudovirions were collected. HeLa cells or CR PKTECs were rinsed with PBS and infected with pseudovirions (Simmons et al. 2004; Wang et al. 2008). Cells were lysated with cell culture lysis reagent and the luciferase activity was read using Luciferase Assay System from Promega (E1500). Relative luciferase units (RLU) were plotted by using cell lysates without pseudovirion infections as negative controls.
-
Publicly usable online RNA sequencing datasets of total RNA from 20 human tissues reported in SRP056969 were used to analyze the level of ACE2 expression. Normalized expression level RPKM (reads per kilobase per million reads) and raw counts were available directly online. Single cell RNA sequencing (scRNA-seq) dataset for kidney was retrieved from www.kidneycellatlas.org or a special website portal (www.covid19cellatlas.org) (Stewart et al. 2019).
Cell Culture
Matrigel Organoids 3D Cultures
Immunofluorescence Staining
Anchorage-Independent Growth Analysis
RNA Extraction and Real Time RT-PCR
Pseudovirus Assay
Acquisition and Analysis of ACE2 Gene Expression from Public Datasets
-
We collected online RNA sequencing datasets and analyzed the level of ACE2 expression in different organ tissues. As shown in Supplementary Figure S1, kidney tissue expresses the highest level of ACE2 mRNA among the 20 organs reported in SRP056969 dataset, and ACE2 mRNA was predominantly expressed in proximal tubule cells (Supplementary Figure S2). Next, we tested whether CR technique could propagate normal kidney epithelial cells. CR technique has been used to efficiently establish long term cell cultures from variety of tissue types without gene manipulation (Liu et al. 2017). As far as we know, there is no report on long term culture of human normal kidney cells. As shown in Fig. 1, typical KPTECs colonies were observed under CR condition (co-cultured with irradiated J2 fibroblast feeder cells in the presence of the ROCK inhibitor Y-27632). KPTECs were observed in 1 day after initial culture, and proliferated rapidly to reach 80%–90% confluence in 4–5 days. KPTECs maintained a normal morphology in the CR system for > 50 passages after one year in culture. The typical cobblestone-like morphology was shown (Fig. 1). When grown in commercial serum-free medium, CR KPTECs senesced after 3–4 passages (data not shown).
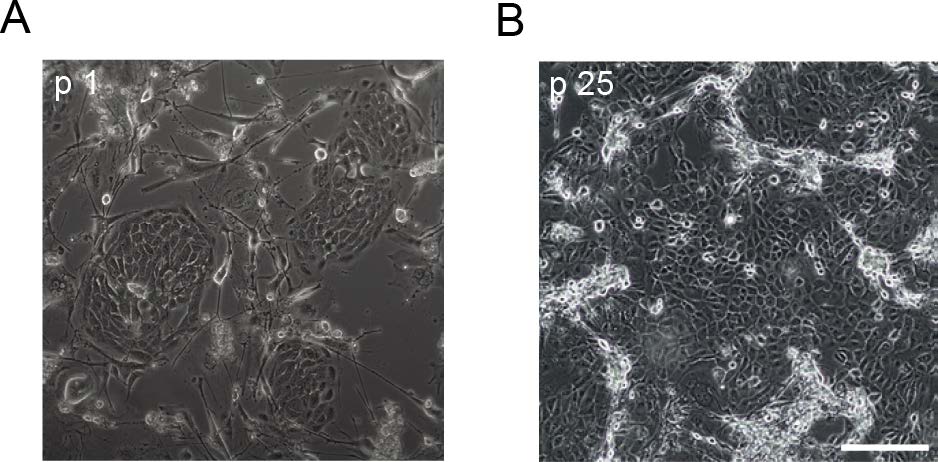
Figure 1. Generation of long term cultures of normal human KPTECs. The normal KPTECs were grown under CR condition (mouse swiss 3T3 cells, J2 clone, and 5 μmol/L Y-27632) as described in Materials and Methods. A Morphology of CR KPTECs at passage 1. B Morphology of CR KPTECs at passage 25. Scale bar, 200 μm.
The differentiation potential of normal cells is important for their physiological function. Normal epithelial cells could be able to maintain their differentiation potential under in vitro and in vivo differentiation conditions, while transformed or malignant cells usually loss their ability to differentiate to functional cells. Previous studies already demonstrated that CR cells from airway, prostate, breast, cervical and skin tissues were able to form well differentiated structures under in vitro organoids or in vivo renal capsule experiments (Suprynowicz et al. 2012; Liu et al. 2017). Matrigel basement membrane matrix is an important regulatory factor to maintain homeostasis of cells and tissue morphogenesis (Lee et al. 2007). Thus, we next investigated whether the later passage (p30) CR cells from normal KPTECs were able to maintain the differentiation properties of normal cells when grown in 3D Matrigel organoid cultures. As shown in Fig. 2A, CR KPTECs formed acinar structures with a well-defined cell/Matrigel interface and showed polarized periphery of the colony in Matrigel organoids.
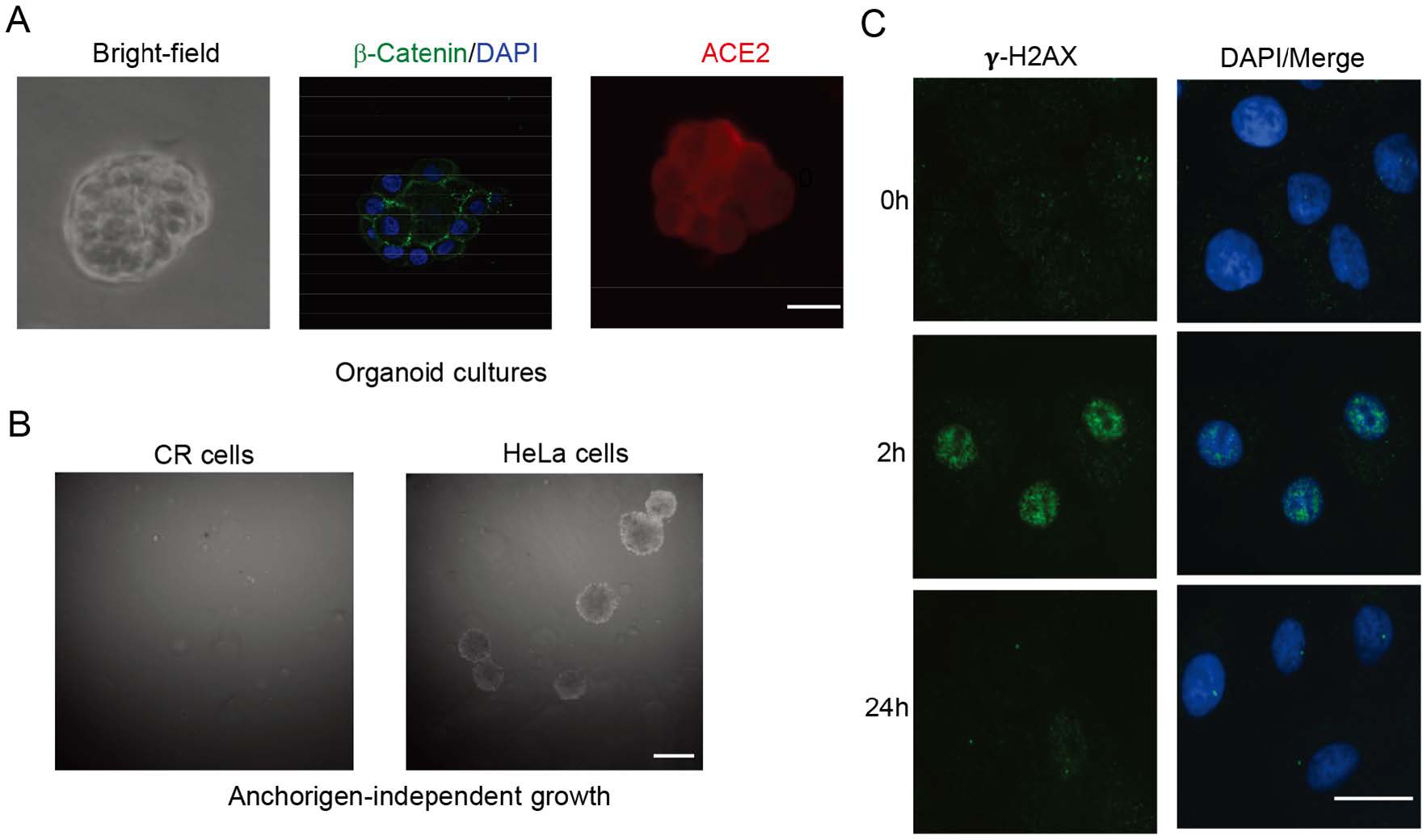
Figure 2. Characterization of CR KPTECs. A Organoid cultures of KPTECs. KPTECs were cultured in Matrigel 3D organoid culture as described in Materials and Methods. Left: bright field image of organoids. Middle: beta-catenin and DAPI. Right: Expression of ACE2. Scale bar, 20 μm. B CR KPTECs did not form colonies in soft agar. CR KPTECs did not form colonies in soft agar, but HeLa cells did. Scale bar, 1000 μm. C CR KPTECs maintained ability to repair DNA damage. Phosphorylated H2AX foci and DNA were stained and images were captured using immunofluorensence microscopy. CR KPTECs were able to repair DNA damage induced by irradiation in 24 h. Scale bar, 10 μm.
Anchorage-independent growth is a hallmark of transformed or malignant cells. The transformed or malignant cells can form colonies when cultured in soft agar, while normal cells are not able to grow under this stringent culture condition. Thus, we tested whether our CR KPTECs were able to form colonies in soft agar. As shown in Fig. 2B, CR KPTECs did not formed colonies in soft agar, indicating that they had no anchorage-independent growth properties as cancer cell line did.
-
It is of special interest to further test if CR KPTECs retain characteristic of original normal tissue at the molecular level. For that purpose, we used immunofluorescence to visualize phosphorylated H2AX foci in CR KPTECs at various times after irradiation (2 h and 24 h). Foci are believed to represent the sites of DNA double-strand break (DSB) and H2AX phosphorylation are the early events in DNA damage response and repair (DDR). After DSBs repair, foci resolution occurs by dephosphorylation of the p-H2AX protein, while persistence of the foci at late time points is thought to represent residual unrepaired DSB. By comparing the formation and resolution of the foci in the cells at the different time points after irradiation, we can determine the DSB repair kinetics and the status of the DDR response. We analyzed the time-dependent change of gamma irradiation induced-H2AX foci in CR KPTECs. The results showed that the DDR response occurred in CR KPTECs at 2 h after irradiation (Fig. 2C). However, almost undetectable fraction of the initial foci persist after 24 h were observed, suggesting that the cells had the ability to repair DNA damage.
-
Since transport functions are unique property of kidney tubule cells, we tested the expression of KPTECs specific sodium-dependent phosphate transporters in CR KPTECs. We performed RT-PCR for detection of SLC34A3 mRNA. Our results demonstrated that CR KPTECs expressed SLC34A3, while HeLa cells did not express SLC34A3 as a negative control (Fig. 3A). Another specific transport system expressed in KPTECs is the megalin/cubilin multiligand receptor, which plays an essential role in endocytotic uptake of filtered proteins by the proximal tubule in kidney. We observed similar pattern for expression of cubilin in CR KPTECs using RT-PCR (Fig. 3A). These results suggested that CR KPTECs maintain lineage function with expression of specific functional genes in the proximal tubules.
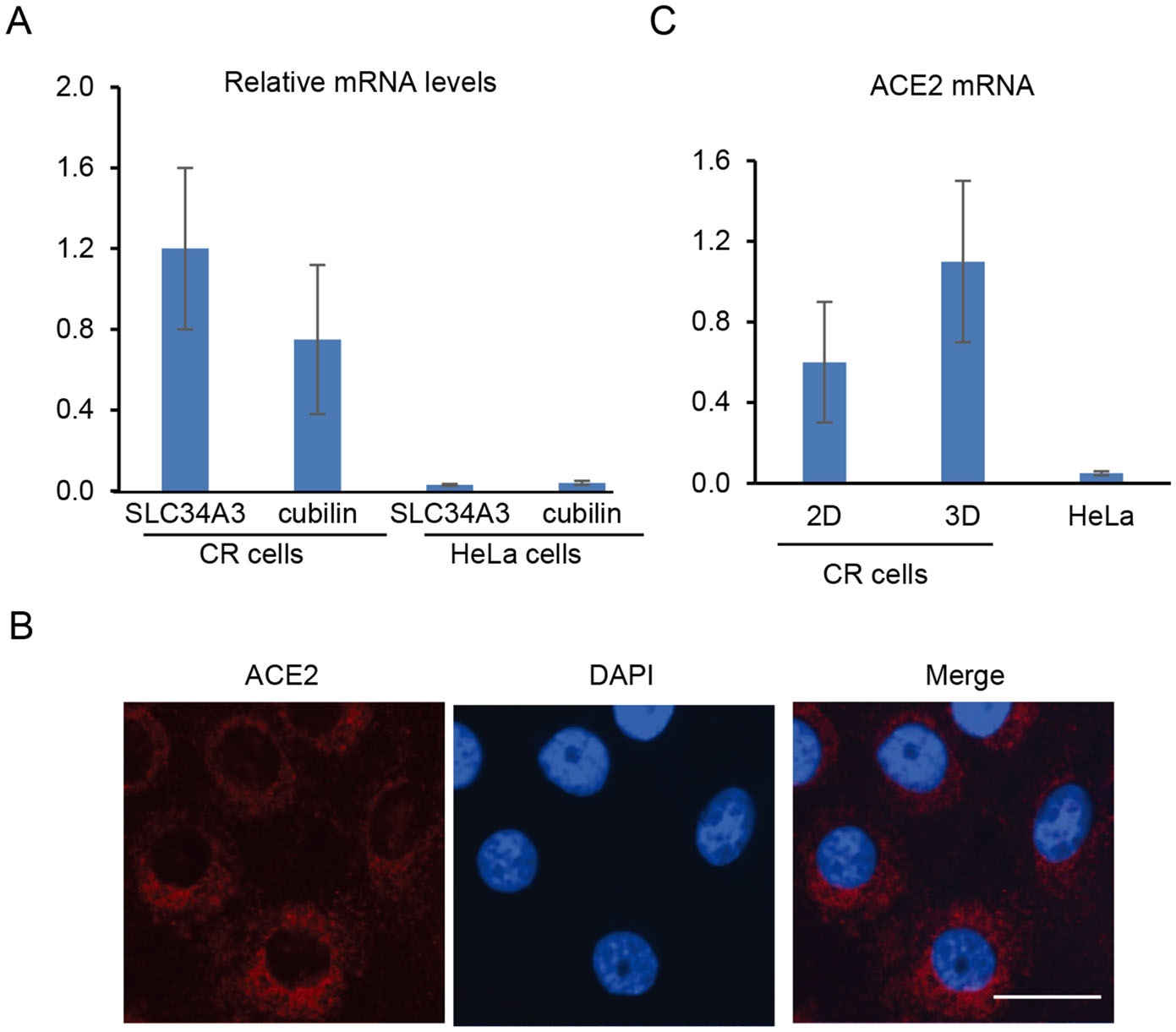
Figure 3. Expression of endogenous transporters and ACE2 in KPTECs. A Expression of endogenous SLC34A3 and cubilin. Expression of two transporters was measured using quantitative RT-PCR. β-Actin was used as an internal control. Both SLC34A3 and cubilin were expressed in CR KPTECs, but not in HeLa cells. Expression of endogenous ACE2 in CR KPTECs was measured using IF (B) and quantitative RT-PCR (C). Scale bar, 10 μm.
-
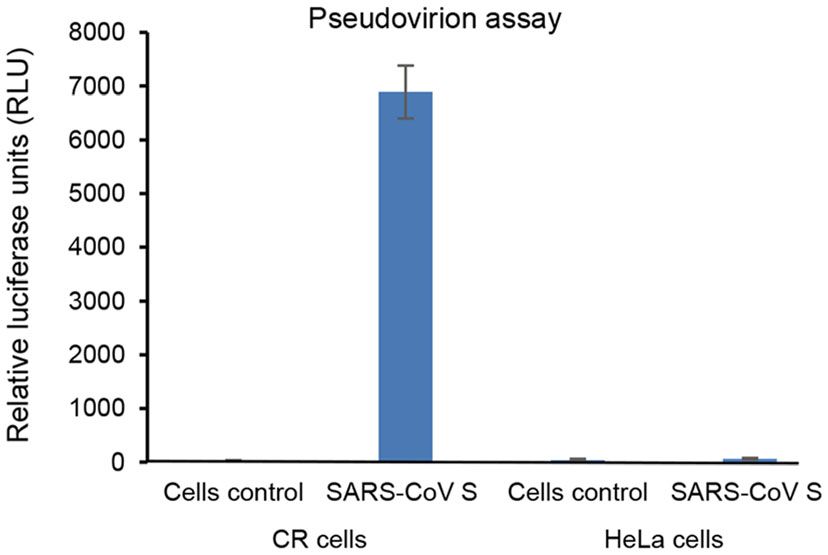
As we described above, some cases with COVID-19 progress to severe pneumonia and multi-organ failure including kidney. To study whether our long term culture system can be used for studies of SARS-CoV-2 and COVID-19, we performed immunofluoresence and RT-PCR to test the expression of ACE2 in 2D and 3D cultures. The ACE2 mRNA and protein were detectable in both 2D and 3D cultures (Figs. 2A, 3B, 3C). Interestingly, our RT-PCR demonstrated expression of endogenous ACE2 in CR KPTECs, not in HeLa cells (Fig. 3C). We also observed a relative higher (around twofold) expression of ACE2 in 3D organoids compared to that in 2D CR conditions (Fig. 3C). Finally, we generated pseudovirions in HEK293T cells transfected with SARS-CoV S plasmid, psPAX, and lenti-Luc. CR KPTECs and HeLa cells were infected by pseudovirions to investigate whether these cells could be infected by SARS-CoV pseudovirions. As shown in Fig. 4, CR KPTECs infected with pseudovirions showed RLUs of 6890 ± 490, while CR KPTECs without pseudovirion infection had RLUs of only 28 ± 8. RLUs were below 100 in HeLa cells with or without SARS-CoV S pseudovirion infection. Thus, CR KPTECs were permissive for SARS-CoV infection. These data suggested that our integrated 2D CR and 3D organoids would be an appropriate physiological human cell system for study of SRAR-CoV and SARS-CoV-2 and their associated kidney injuries, especially for virus entry to KPTECs, innate immune response of host cells, drug screening for anti-SARS-CoV-2 and safety evaluation of kidney functions.
Genaration of Long Term Culture of Normal Human KPTECs
CR KPTECs Retain Their Ability to Repair DNA Damage
CR KPTECs Express Functional Specific Transporters
CR KPTECs Express ACE2 Which is a Receptor for SARS-CoV and SARS-CoV-2 Infections
-
In this study, we firstly established long term cultures of human KPTECs using CR technology. These CR cells maintained their differentiation potential and the ability to repair DNA damage induced by irradiation. These cells also expressed lineage functional markers of kidney proximal tubule epithelial cells, including SLC34A3 and megalin/cubilin. These data suggested that CR KPTECs would be used as an ex vivo model for studies of kidney diseases or kidney injury associated with other systemic diseases (e.g., diabetes), and discovery of novel biomarkers and targets.
As we discussed above, mortality of severe patients with COVID-19 are relative high due to preexisting conditions and multi-organ failure (Wang T et al. 2020; Wang Z et al. 2020; Wu and McGoogan 2020; Xu L et al. 2020; Xu Z et al. 2020; Yang X et al. 2020; Yao et al. 2020; Ye et al. 2020; Zhang H et al. 2020). As SARS-CoV-2 enter host cells via ACE2 protein receptor, it is highly possible that SARS-CoV-2 infect all ACE2 expressing cell types. It was reported that ACE2 is highly expressed in intestine, testis, kidney, heart, etc., and this might help to explain multi-organ injury (Zou X et al. 2020). In particular, a number of studies have reported kidney injury in severe COVID-19 cases (Huang et al. 2020; Li et al. 2020). Furthermore, culturing SARS-CoV-2 virus and SARS-CoV-2 mRNA were detected from urine samples of COVID-19 patients (Ling et al. 2020; Rothe et al. 2020; Xie et al. 2020; Young et al. 2020), indicating the possible replication of SARS-CoV-2 in urological system, especially in kidney and bladder. Thus, it would be important to study the possibility of SARS-CoV-2 infection with urological system.
In terms of the questions above, we need model systems to study infection of SARS-CoVs in ACE2 expressing cell types, especially in kidney epithelial cells (Hamming et al. 2004; Zou X et al. 2020). Transgenic mice with human ACE2 (Yang et al. 2007; Netland et al. 2008) would be used for modeling disease in vivo, however, it's hard to mimic human diseases due to species-related restriction conditions. Animal models are usually very expensive and need much longer time to complete, thus they cannot be used for any high throughput screening. Another option is human cell lines. Cancer cell lines usually gain the ability to proliferate in very stringent conditions, such as soft agar, and they loss their specific differentiation potential. Thus, those cancer cell lines are not suitable to study "normal" or "physiological" response of host cells to viral infections, although viruses may lead lytic infections in some cancer cell lines. Therefore, much efforts have been made for generating immortalized cell lines (Ryan et al. 1994; Racusen et al. 1997; Kowolik et al. 2004; Orosz et al. 2004; Wieser et al. 2008). As we described above, HPV16 E6/E7, SV40, and hTERT have been widely used for immortalization of human primary cells (Ryan et al. 1994; Racusen et al. 1997; Kowolik et al. 2004; Orosz et al. 2004; Wieser et al. 2008). These cell lines have been widely used in many study fields, including cell biology, production of research reagents and modeling human diseases. However, due to pathways altered by viral or cellular genes or integration of viral/cellular gene to host genome, these immortalized cell lines cannot reflect the physiological status of normal human cells. A related study reported the susceptibilities of a panel of cell lines to SARS-CoV (Kaye 2006). Among the 21 cell lines, including several cell lines from monkey and other species, only three human cell lines (HEK293, Hep G2 and Huh-7) are susceptible (Kaye 2006). Besides, SARS-CoV can achieve high viral load in some cell lines without specific CPE (cytopathic effect) (Kaye 2006). However, none of these cell lines retain physiological functions.
A recent study demonstrated that active SARS-CoV-2 replication was detected in tissues of the upper respiratory tract, especially virus load and shedding were very high during the first week of symptoms (Wolfel et al. 2020). This suggests an urgent need to establish normal airway cells model from upper (nasal cavity) to lower (lung) respiratory tract to study interactions between SARS-CoV-2 and host epithelial cells. CR has been widely used for generation of various patient-derived cell models that can be used for modeling human disease, drug discovery and tissue regeneration (Borodovsky et al. 2017; Wolf et al. 2017; Yuan et al. 2017; Alkhilaiwi et al. 2018; Brewington et al. 2018; Jin et al. 2018; Moorefield et al. 2018; Peters-Hall et al. 2018; Sayej et al. 2018; Wang et al. 2018; Zhang et al. 2018; Alkhilaiwi et al. 2019; Nicolas et al. 2019; Palechor-Ceron et al. 2019; Wan 2019; Wang et al. 2019; Chai et al. 2020; Krawczyk et al. 2020; Peters-Hall et al. 2020; Wang Y et al. 2020). In the current study, we established long term cell cultures of KPTECs using CR technology. Our CR KPTECs maintain lineage differentiation potential and express cell type-specific function markers and ACE2 protein. These cells can serve as a physiological cell model for the study of SARS-CoVs and kidney functions, innate immune response of kidney cells to viruses and a novel platform for drug discovery and safety evaluation.
-
This work was supported by the National Natural Science Foundation of China (81571396 and 81771528); Science, Technology and Innovation Commission of Shenzhen Municipality (JCYJ20170411090932146, JCYJ20170818110544730).
-
HL: Conceptualization, SX, MW, SC, TZ, LY, JL: Laboratory work and data analysis. SX, MW, HL: Drafting manuscript. All authors have read and approved the submitted version of manuscript.
-
The authors declare no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.

















 DownLoad:
DownLoad: