HTML
-
Alphavirus is a group of enveloped single-stranded positive-sense RNA viruses, which currently includes 31 species and is classified into 8 complexes according to their antigenicity (Forrester et al. 2012; Markoff 2015). In nature, most alphaviruses circulate between invertebrate insect vectors and vertebrate hosts. The main insect vectors are mosquitoes, and the vertebrate hosts are generally birds or mammals (Lim et al. 2018). Many alphaviruses cause serious but not life-threatening diseases (Marks and Marks 2016; Taylor et al. 2015; Weaver and Paessler 2009). With in-depth studies on the structures and functions of alphavirus proteins, some alphaviruses, such as Semliki Forest virus (SFV) and Sindbis virus (SINV), have been widely explored as vectors for vaccines and as oncolytic viruses (Lundstrom 2014, 2016, 2017).
Currently, oncolytic viruses are emerging in a powerful anticancer strategy in which natural or genetically modified viruses selectively kill tumor cells while remaining harmless to normal cells. In addition to direct oncolysis, oncolytic viruses can release pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) to activate the antitumor immune response (Raja et al. 2018). In 2015, the US Food and Drug Administration and the European Medicines Agency approved talimogene laherparepvec (known as T-VEC) for the treatment of melanoma patients (Eissa et al. 2018; Pol et al. 2016). Moreover, the combination of T-VEC and anti-PD-1 antibody Keytruda has led to a more powerful therapeutic effect than either compound alone. The overall response rate was 62%, and the complete response rate was 33%, suggesting the great potential of the strategy of combining an oncolytic virus with an immune checkpoint inhibitor (Ribas et al. 2017).
The M1 virus is a strain of alphavirus that was isolated from Culicine mosquitoes collected on Hainan Island in China. A previous study characterized alphavirus M1 as a novel natural oncolytic virus that selectively targets and kills ZAP-defective human cancers by causing irreversible endoplasmic reticulum (ER) stress-induced apoptosis (Lin et al. 2014). Furthermore, M1 virus is not pathogenic in nonhuman primates, as indicated after multiple rounds of repeated intravenous injections (Zhang et al. 2016), and a series of small-molecule drugs significantly enhanced the M1 virus killing of tumor cells (Cai et al. 2017; Tan et al. 2018; Zhang et al. 2017). High safety and efficiency make M1 virus a potential antitumor candidate for drug preclinical development. However, the basic life cycle of M1 virus in host cells remains unknown. It is important to study how it infects, replicates and assembles progeny viruses and how the virus particles interact with the host during replication, as these findings may support the development of M1 virus as an oncolytic drug.
Here, we used transmission electron microscopy (TEM) to visualize the main steps of the life cycle during M1 virus infection and the virus-induced cytopathic effect on liver cancer cells, which is the most frequent fatal malignancy (Anwanwan et al. 2020). By using the M1 virus-sensitive liver cancer HepG2 cell line, we found that M1 virus induced vacuolization of cancer cells during infection and karyopyknosis, a typical apoptosis indicator. The life cycle of the virus, notably attachment and entry, precursor formation, maturation and progeny release were also observed. In particular, this investigation has also made some novel discoveries, suggesting that the ER may be involved in nucleocapsid assembly, and nucleocapsids may obtain envelopes in the CPV-II inside cells. These findings clearly show the process of M1 virus infection, replication and maturation in HepG2 cells and reveal the interactions between M1 and HepG2 cells.
-
Cell lines were purchased from American Type Culture Collection and cultured in DMEM, MEM or RPMI-1640 medium supplemented with 10% (vol/vol) FBS and 1% penicillin/streptomycin (Life Technologies, USA).
-
For better observation, we used recombinant virus rM1-GFP, which was reconstructed based on the natural M1 virus genome sequence. The GFP gene was inserted between protein CP and PE2 of M1 genome, and protease cleavage sites were designed at both ends of GFP. When M1 virus structural proteins are translated, the GFP protein would be cleaved automatically without affecting the structure of M1 virus. rM1-GFP virus was grown in Vero cells, and the virus titer was determined by TCID50 assay using BHK-21 cells. A high viral titer was obtained by ultracentrifugation as follows: 250 mL of the virus supernatant was collected and centrifuged at 5000 × g (SX4750) and 4 ℃ for 10 min to remove cells and large fragments in the supernatant and then filtered through 0.22 μm syringedriven filters to remove proteins of large molecular weights. The virus filtrate was centrifuged at 80, 000 × g (SW 32 Ti) and 4 ℃ for 1 h, and the supernatant was decanted. The virus was dissolved in 25 mL of VP-SFM (a special culture medium for virus). After determining the high virus titer, the samples were stored at - 80 ℃.
-
For virus entry, 8 × 105 HepG2 cells were seeded in 60 mm petri dishes with 4 mL of media. After the cells adhered to the wall, they were infected with rM1-GFP virus at 100 MOI for 15 min to 2 h.
To determine the virus life cycle, 8 × 105 HepG2 cells were seeded in 60 mm petri dishes with 4 mL of media. After adhering to the wall, the cells were infected with rM1-GFP virus at 3 MOI for 6 h, 12 h and 18 h.
-
The treated cells were washed with PBS, fixed with 2.5% glutaraldehyde at 4 ℃ overnight, and washed three times with PBS. Cell pellets were collected and subjected to osmium tetroxide at 4 ℃ for 1 h and then washed three times with PBS. Then, the pellets were dehydrated in an ascending ethanol series, embedded in Epon and cut into ultrathin sections. The sections were collected on naked grids and stained with uranyl acetate and lead citrate. The grids were examined with a JOEL JEM-1400Flash electron microscope operating at an acceleration voltage of 120 kV. Finally, the images were recorded with a CCD camera (Morada G3, Emsis).
-
TEM images of the M1 particles at different infection stages and locations were collected at a nominal magnification of 10 k with a Gatan Rio CMOS camera. The final pixel size was 0.65 nm. The contrast of each image was first inverted by e2proc2d.py because the software used for the assay recognizes the density as white, and e2boxer.py in EMAN2 (Tang et al. 2007) was used to boxed out and select the particles in different areas. Relion 2.0 (Kimanius et al. 2016) was used to perform reference-free 2-dimensional (2D) classification. The particles were distinguished automatically into 5 classes based on their features, except for the particles inside the CPV-Ⅱ-2 because of the small number of particles. To determine the precise sizes of each group of particles, the weighted average method was applied to calculate the average diameters. Specifically, the ratio of the numbers in each classification was used as the weight coefficient (W), and the weighted average diameter of the particles in this group is Dw = W1*D1 + W2*D2… + W5*D5. The summary of W1–W5 is 100%. D represents the diameter of a particle in a classification.
-
HepG2 cells (4 × 104) were seeded in 24-well plates with 500 μL of medium per well. After adhering to the wall, the cells were treated with rM1-GFP at 0.001 MOI, 0.01 MOI, 0.1 MOI, 1 MOI and 10 MOI for 72 h. Then, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-tetrazolium bromide (MTT, MP Biomedicals, USA) was added to the cells (1 mg/mL final concentration), and the cells were allowed to grow at 37 ℃ for another 3 h. The MTTcontaining medium was removed, and the MTT precipitate was dissolved in 1 mL of DMSO. The optical absorbance was determined at 570 nm using a microplate reader.
-
HepG2 cells (2.4 × 105) were seeded in 6-well plates with 2 mL of medium per well. After adhering to the wall, the cells were treated with rM1-GFP at 0.001 MOI, and the cell culture supernatant was collected 0 h, 24 h, 48 h and 72 h after infection. The virus titers were determined by TCID50 assay using BHK-21 cells.
Cell Culture
Virus
Transmission Electron Microscopy Preparation and Observations
Transmission Electron Microscopy Analysis
2D Reference-Free Classification Assay
MTT Assay
Replication Efficiency Assay
-
To establish a suitable cell model for studying the life cycle of M1 virus, we chose the HepG2 hepatic carcinoma cell line. In order to better observe the infection of M1 virus, we used the recombinant rM1-GFP virus, whose genome was inserted GFP gene and maintained the characteristic of M1 virus. We first tested the killing efficiency of rM1-GFP virus in the HepG2 cells and found that infection with rM1-GFP for 72 h, even at the lowest dose of rM1-GFP (0.001 MOI), reached a killing efficiency of 95%, meaning that rM1-GFP virus induced substantial cytopathic effects in the HepG2 cells (Supplementary Fig. S1A). In addition, we tested the replication efficiency of rM1-GFP in HepG2 cells and found that rM1-GFP virus replicated quickly and was released into the culture supernatant in large quantities (Supplementary Fig. S1B). We concluded that the HepG2 cells were highly sensitive to rM1-GFP virus, and prepared EM samples by infecting HepG2 cells with a moderate dose of rM1-GFP virus (MOI = 3) to observe rM1-GFP life cycle and cytopathic effect induced by rM1-GFP virus. We designated samples with an infection time less than or equal to 6 h as the early stage in the rM1-GFP life cycle and samples with an infection time longer than or equal to 12 h as the late stage, corresponding to rM1-GFP virus maturation in and release from HepG2 cells.
Compared with that in the uninfected cells, after 6 h infection, the endoplasmic reticulum (ER) showed slight distension in the infected cells (Fig. 1A, 1B), and with an increase in infection time, the levels of ER swelling became more serious (Fig. 1C). Moreover, the infected HepG2 cells showed other typical apoptotic characteristics, such as the condensation and marginalization of nuclear chromatin (Fig. 1D). However, no apoptotic body was observed, even though the cells showed catastrophic levels of vacuolization. Plasma membranes still remained intact, but pseudopodia extending from cells were dramatically reduced and shortened, with severe vacuolization and no cytoplasm inside them (Fig. 1E). In addition, morphological changes of mitochondria in the virus-infected cells were also prevalent. The mitochondrial inner membrane and ridges were damaged, resulting in vacuolization (Fig. 1F). These data suggest that rM1-GFP virus infection induces the apoptosis of HepG2 cells.
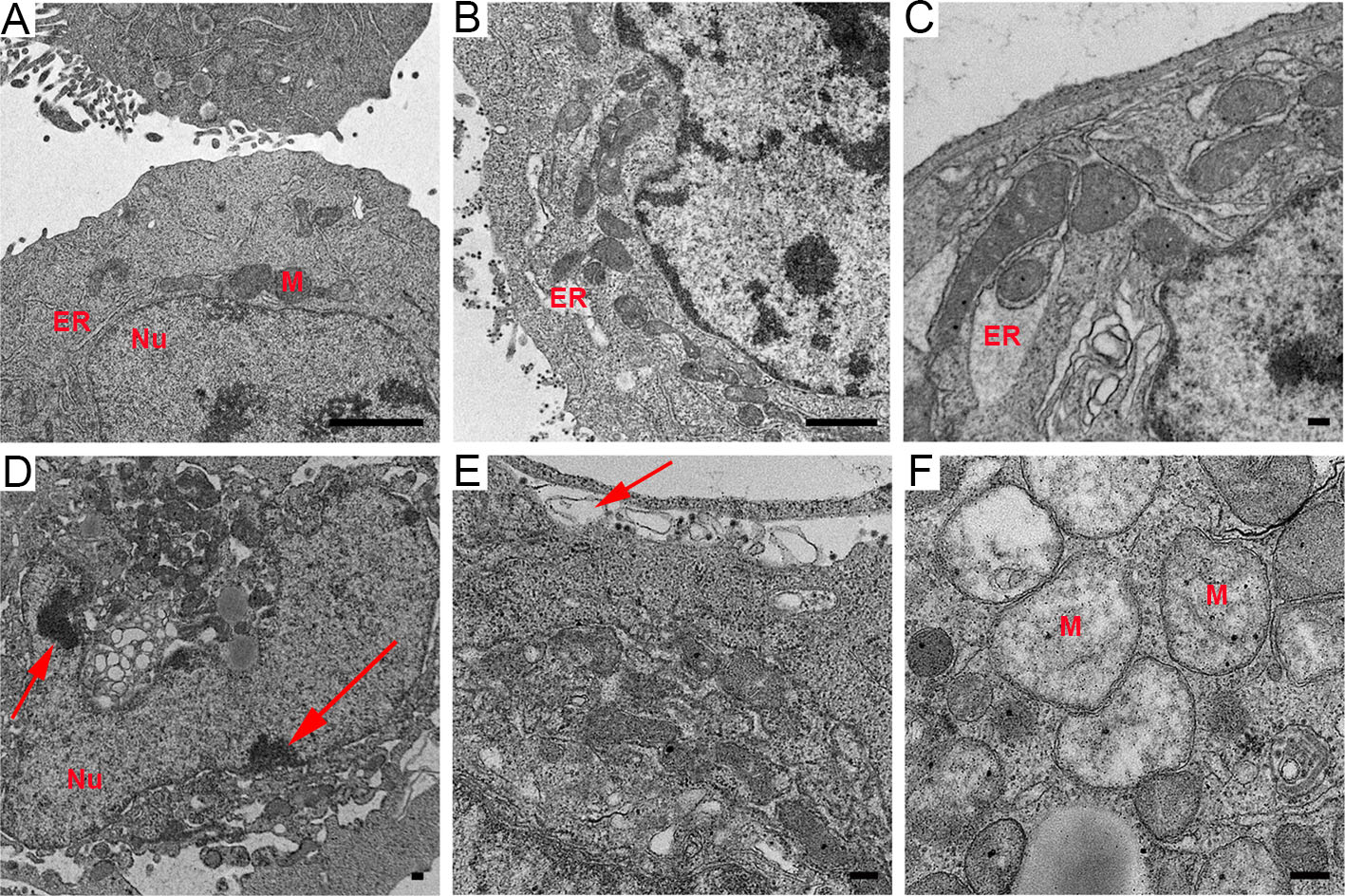
Figure 1. M1 virus replicated and induced apoptosis in the HepG2 liver cancer cell line. A Control HepG2 cells uninfected with M1 virus. B ER distension. C ER became swollen. D Condensation and marginalization of nuclear chromatin are indicated by red arrows. E The decrease and vacuolization of pseudopodia are indicated by red arrows. F Vacuolization of mitochondria with damage to the membrane and ridges. ER: endoplasmic reticulum. M: mitochondria. Nu: nucleus. Time points: 6 h (B), 12 h (C) and 18 h (D–F). Bar: 2 μm (A, B) and 200 nm (C–F).
-
To visualize the initial events of rM1-GFP virus infection in substantial detail, HepG2 cells cultured at 37 ℃ were also inoculated with a high dose of rM1-GFP (MOI = 100), fixed at 15 min ~ 2 h after infection, and observed with EM. The results showed that intact ''big'' rM1-GFP virus particles consisted of a small core of high electron density with a halo-like structure surrounding the core (Fig. 2), an observation consistent with that of other reported alphaviruses (Zhang et al. 2011). Many studies have reported that viruses may be internalized through a variety of endocytic processes, including macropinocytosis, clathrinmediated endocytosis, caveolae, and clathrin- and caveolinindependent mechanisms (Cossart and Helenius 2014; DeTulleo and Kirchhausen 1998). Our results showed that virus particles aggregated and appeared in an invaginated space of the cell membrane, which may be indicative of the process of a virus entering the cell through the endocytosis pathway but not micropinocytosis (Fig. 2A). In the 2 h p.i., a group of ''large'' particles with halo-like structures accumulated in several vesicles inside the cell, suggesting that multiple virus particles may synchronously invade cells through one endocytosed vesicle (Fig. 2B). In addition, we also found a group of ''small'' particles with high electron density in the cytoplasm matrix rather than encapsulated in vesicles (Fig. 2B), which may be nucleocapsids released into the cytoplasm after their viral envelopes were removed in the endosome.
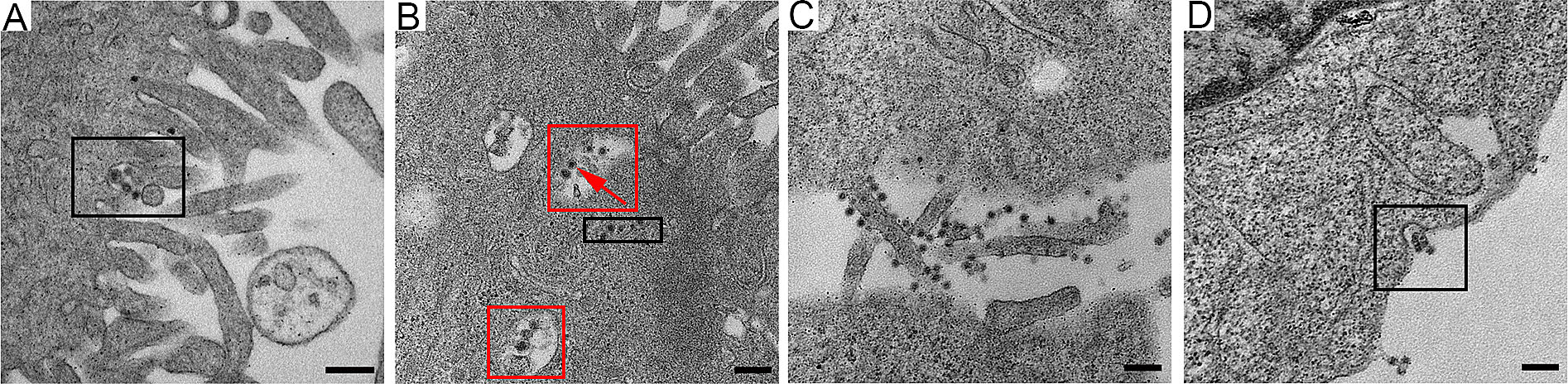
Figure 2. The early stage of M1 virus infection of the HepG2 cancer cells. A A few virus particles aggregated near the pseudopodia and appeared in the invaginated space (the circled area) of the cell membrane. BSeveral M1 viruses gathered in the same vesicle inside the cell (circled with red), and nucleocapsids (with only a high electron density core) appeared in the cytoplasm matrix (circled with black). C Many viruses absorbed on the cellular pseudopodia. D M1 virus particles also gathered in pits in the cell membrane. Time points: 15 min (A), 2 h (B) and 6 h (C, D). MOI: 100 pfu per cell (A, B) and 3 pfu per cell (C, D). Bar: 200 nm.
To prevent the formation of artificial features caused by high-dose infection, we also observed the early stage of M1 infection in the sample of cells infected with a moderate dose of rM1-GFP virus (MOI = 3) and fixed 6 h p.i. Consistently, many virus particles were found attached to cellular pseudopodia, and few particles were attached to the main body of the cells (Fig. 2C). We also found that several virus particles accumulating in pits of the cell membrane (Fig. 2D), similar to those we observed in the samples of cells infected with a high dose of rM1-GFP virus (Fig. 2A). Taken together, our data show that rM1-GFP virus may enter cells via endocytosis and that more than one viral particle can enter simultaneously through one endocytosed vesicle, which may promote highly efficient infection.
-
After viral infection, viral envelope fuses with the endosome membrane, nucleocapsid is released into the cytoplasm, and viral RNA begins to translate into the nonstructural proteins necessary for replication (Kääriäinen et al. 1975). In both early and late stages of rM1-GFP virus infection, we found structures of plasma membrane invagination called ''spherules'' (Fig. 3A), which are generally believed to form a favorable microenvironment for RNA synthesis (Kallio et al. 2016). And the cytopathic vacuoles type I (CPV-I), which are characterized in structures of regularly spaced ''spherules'' that projected from their interior surfaces, were also observed in the sample of 12 h p.i. (Fig. 3B) (Grimley et al. 1968). After the synthesis of viral genomes and subgenomes in these spherules, structural proteins are translated into the cytoplasm, and nucleocapsids begin to assemble. Surprisingly, in addition to finding nucleocapsids in the cytoplasm matrix (Fig. 3B), some nucleocapsid-like particles with high electron density were also found inside the ER cavity of the cells in all samples of 6 h, 12 h and 18 h p.i., which were probably newly synthesized nucleocapsids consisting of capsid proteins and viral RNA, also known as viral precursors (Fig. 3C, 3D). These results indicate that the ER might participate in the process of nucleocapsid assembly.
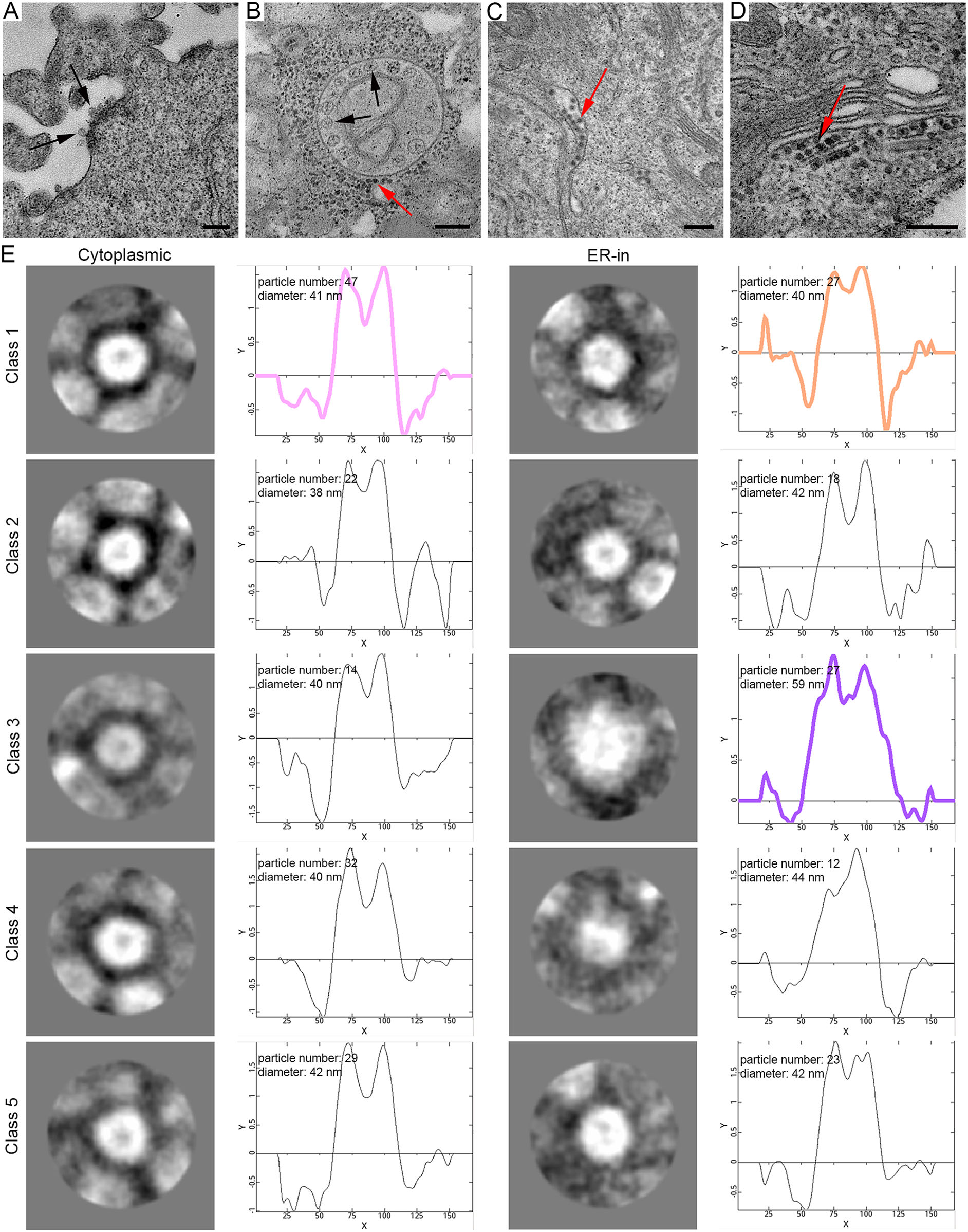
Figure 3. M1 virus precursors were scattered in the cytoplasmic matrix and inside the ER cavity. A ''Spherules'' were formed on the plasma membrane (indicated by black arrows). B Representative morphology of the CPV-I and ''spherules'' were formed inside it. Nucleocapsids were distributed in the cytoplasm (indicated by the red arrow). C and D Virus particles with a high electron density core were scattered inside the ER cavity. E The representative 2D classification of particles in the cytoplasmic matrix and inside the ER, with the number, average diameter and radial density curve shown. The contrast of the images was inverted because the software recognizes the density as white. The colored curves correspond to the radial density curves of the selected classifications. Time points: 6 h (A and C), 6 h (B) and 18 h (D). Bar: 200 nm.
To investigate whether the particles scattered in the ER cavity were nucleocapsids, we used the single-particle analysis method to compare the sizes and structures of the particles inside the ER and cytoplasmic matrix of the sample cells. We manually set up 5 classes, the software would automatically sort the selected particles into 5 classes, according to their features including different diameters, shapes, etc. (Fig. 3E). The results demonstrated that all the particles were nearly round and had very close size. However, carefully checking these 5 classes, we also found they were very slightly different in shape, for example, particles in class 1 were rounder than those in class 5. All these slight ''difference'' in shape may be caused by the sample preparation for TEM, such as chemical fixation, ultrathin section, metal staining, etc. Therefore, we only focused on the size difference of these particles. The average diameter of the particles in the cytoplasmic matrix, which consisted only of concentrated cores, were 38 ~ 42 nm in agreement with the reported diameter of nucleocapsid. In addition, inside the ER, approximately 74.7% (80/107) of the particles had a diameter of approximately 40 nm, which was similar to those in the cytoplasmic matrix. Interestingly, we found another group of particles in the ER cavity, accounting for approximately 25.2% (27/107) of those we analyzed, and they had an average diameter of 59 nm, indicating that these virus particles may have been developed in the ER. These results suggested that the ER was possibly involved not only in the assembly of the nucleocapsid but also in the maturation of rM1-GFP virus.
-
After observing viral nucleocapsid assembly, we looked for the appearance of rM1-GFP virus maturation in and release from the cells in the samples of 12 h p.i. and 18 h p.i.. A large number of ''small'' particles, with high electron density, were commonly located in the cytosol, and multiple layers of them were also arranged near the cytoplasmic side of the cell membrane (Fig. 4A), as well as the pseudopodium of one cell (Fig. 4B). Outside and along the plasma membrane, many ''big'' particles with light halos and ''dark'' centers were observed (Fig. 4A). As described above, the 2D classification results demonstrated that the particles inside the cytoplasm were approximately 40 nm (Fig. 3C), and the particles outside of the cells were 65 nm in diameter (Fig. 4C). Therefore, all of these evidences indicated that the particles outside the plasma membrane in the later stage of infection were mature rM1-GFP progeny virus budding and being released to the extracellular environment. These data indicated that the nucleocapsids were packed with envelope proteins, matured and were released by budding from the cell surface.
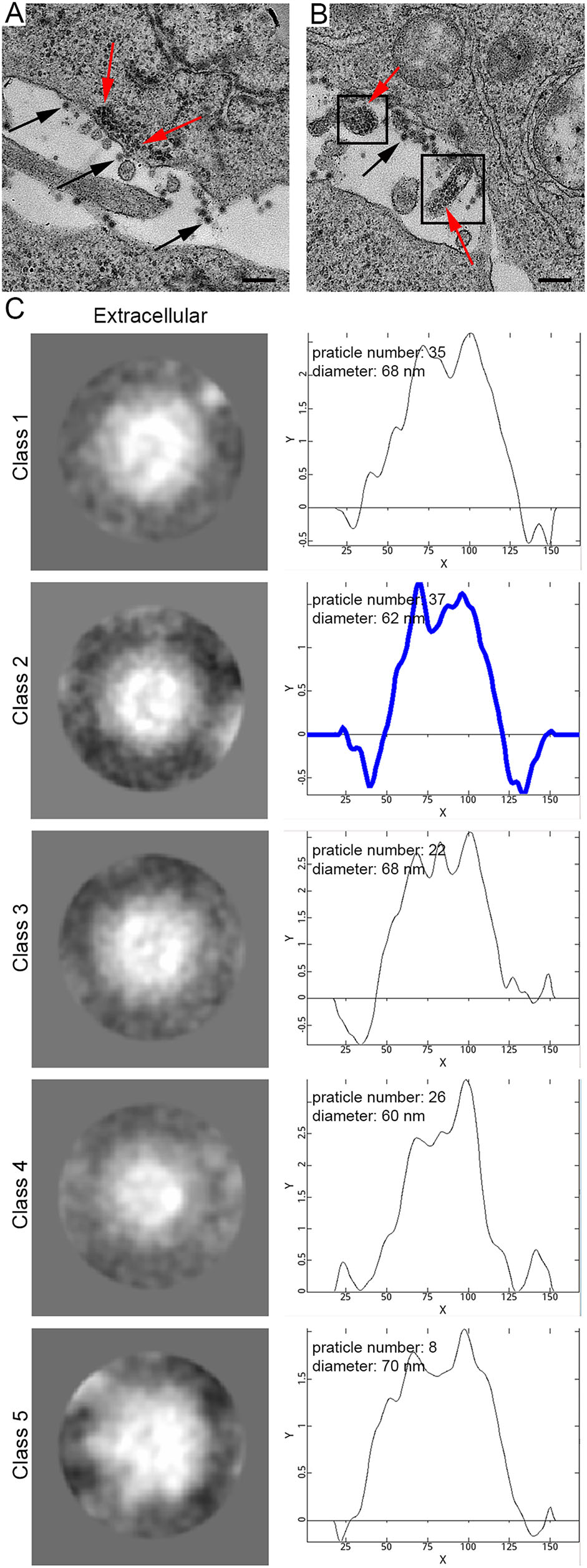
Figure 4. The maturation and release of M1 virus by budding on the plasma membrane. A M1 virus precursors stacked in the cytoplasm near the plasma membrane for budding. Red arrows indicate precursors, and black arrows indicate mature virions. B M1 virus precursors accumulated in the cellular pseudopodia. C The 2D classification of the extracellular particles. The number of particles in the classes, average diameters and radial density curves is indicated. The highlighted blue curve corresponds to the radial density of the selected majority classification. Time point: 18 h. Bar: 200 nm.
-
With increasing infection time, rM1-GFP virus induced severe vacuolization of the HepG2 cells. Several types of cytopathic vacuoles formed in the cytoplasm during the process of rM1-GFP virus replication. Few small vacuoles were scattered in the cytoplasm in the early stages of infection, but they were gradually increased in size and number (Fig. 5A–5C and Supplementary Fig. S2). In addition, masses of virus particles were more commonly accumulated inside and outside vacuoles (Fig. 5D–5F). Two kinds of typical cytopathic vacuoles type II (CPV-II), ''type II-1 (CPV-II-1)'' and ''type II-2 (CPV-II-2)'', were clearly observed in the late stage for virus maturation. Furthermore, we also observed another kind of vacuole, which we named the type-0 cytopathic vacuoles (CPV-0).
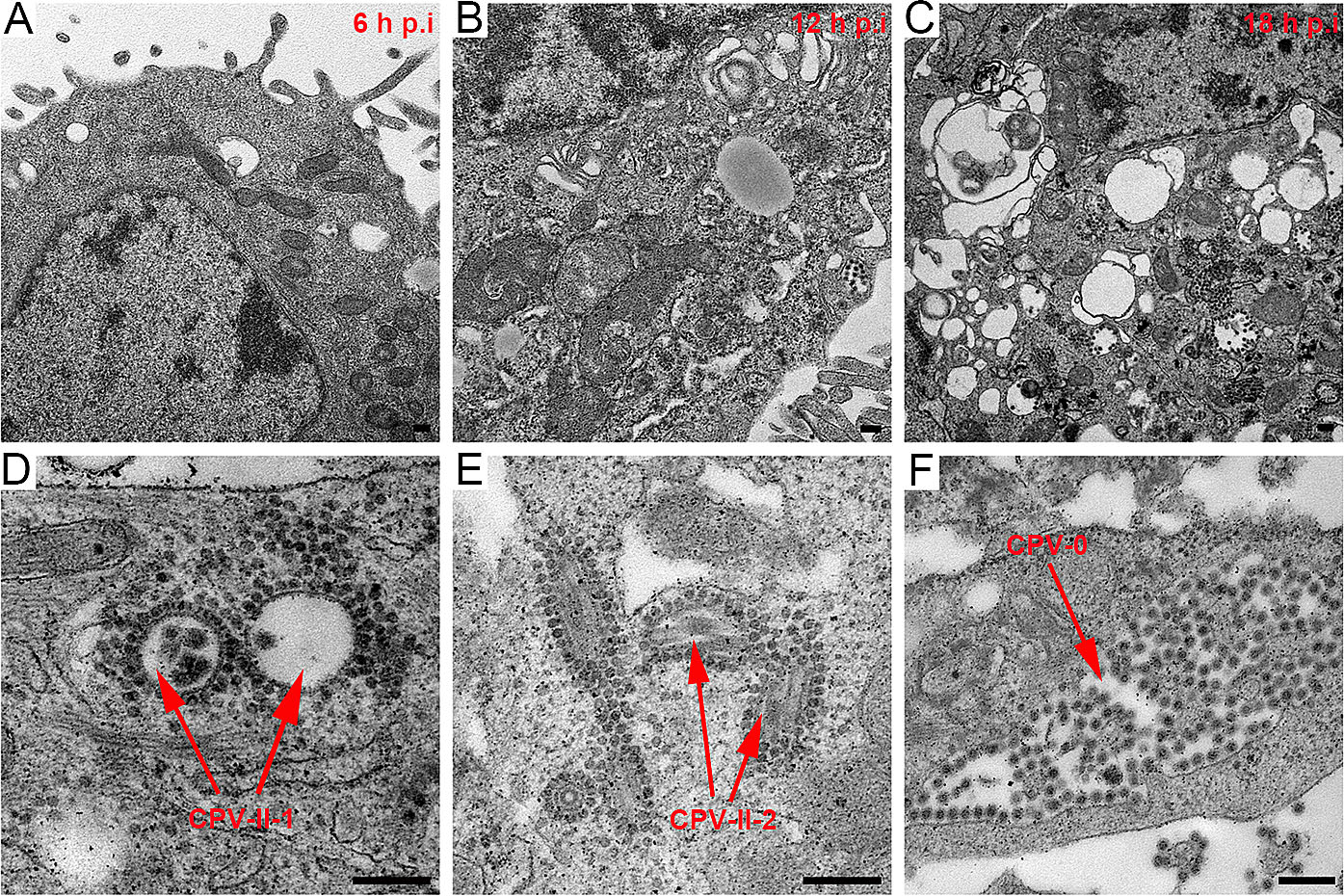
Figure 5. M1 virus induced vacuolization of the HepG2 cancer cells. A–C M1 induced vacuolization of the HepG2 cells in the samples at 6 h, 12 h and 18 h p.i. With increasing infection time, both the number and size of the vacuoles gradually increased in the infected HepG2 cells. D Representative morphology of the CPV-II-1. E Representative morphology of the CPV-II-2. F Representative morphology of the CPV-0. Bar: 200 nm.
CPV-II-1, often appearing in groups, is usually a nearly circular vacuole with a diameter of several hundred nanometers characterized by one or more layers of rM1-GFP virus particles surrounding the cytoplasmic side of the vacuole membrane and existing inside the vacuoles (Fig. 5D). CPV-II-1 first appeared in the samples 12 h p.i. and were abundant in 18 h p.i.. CPV-II-2 consists of a spindle-shaped lumen with one or two internal tubular structures. The length of each tubular structure in the CPVII-2 differed, but the width of each was approximately 30 nm (Fig. 5E). CPV-II-2 was previously found in the sample of 6 h p.i.. The number of both kind of type II vacuoles continually increased in the late stage of infection. CPV-0, in contrast to CPV-I or CPV-II-1/2, had a large number of particles aggregated inside them, but no particles adhered to the outer membrane structure (Fig. 5F). The sizes of the CPV-0 varied on a large scale and appeared in the samples 12 h p.i. and later.
Along with vacuolization of the HepG2 cells, a large number of rM1-GFP progeny virus particles were found to adhere not only to the inner but also to the outer membrane structures of the vacuoles in the samples of 12 h and 18 h p.i. (Fig. 5D, 5E). To distinguish the virus particles more accurately and understand the maturation pathway of rM1-GFP virus, we used the single-particle analysis method to confirm the sizes and structures of these particles at different locations. Each group of virus particles (including the particles inside and outside the CPV-II-1 and CPV-II-2, and inside the CPV-0) was also sorted into five classifications (Supplementary Fig. S3). The weighted average diameter of the particles inside the CPV-II-1, CPV-II-2 and CPV-0 was 61 nm, 60 nm and 64 nm, respectively, similar to that of the mature virions outside the cells, with an average diameter of 65 nm vacuole type. Almost all of the 2D structures had a ''halo'' structure encircling a concentrated core, which corresponded to the ''big'' particles (Fig. 6). Therefore, it is reasonable to consider that the particles inside the CPV-II-1, CPV-II-2 and CPV-0 were mature rM1-GFP virus progeny. The average diameter of the particles outside the CPV-II-1, CPV-II-2 and those dispersed in the cytoplasm was 40 nm, 41 nm and 40 nm, respectively. The 2D viral structures had concentrated cores, which corresponded to the ''small'' particles (Fig. 6). These features were sufficient to show that the particles outside the CPV-II-1 and CPV-II-2 were viral precursors. In conclusion, our results show that, in addition to budding from the plasma membrane, precursors of rM1-GFP virus may also obtain an envelope by budding from CPV-II-1 and CPV-II-2 inside the infected cells, where they mature. Moreover, the mature rM1-GFP particles aggregated inside the CPV-0 (Fig. 5F and Fig. 6), and some of CPV-0 were close to the plasma membrane. These features provide clues that CPV-0 may be critical for releasing mature progeny rM1-GFP virus particles to the outside of the cell through membrane fusion.
M1 Virus Replicates and Induces the Apoptosis of HepG2 Liver Cancer Cells
Virus Attachment and Entry
Assembly of Viral Nucleocapsids
The Maturation and Release of M1 Virus by Budding on the Plasma Membrane
M1 Virus Maturation in and Release from Vacuoles
-
Our study systematically revealed the life cycle of M1 virus in the M1-sensitive HepG2 hepatocellular carcinoma cell line to further understand how M1 virus infects, assembles, matures and releases progeny (Fig. 7). First, M1 virus enters HepG2 cells via endocytosis, and an endocytosed vesicle may contain a plurality of the virus particles. Second, the assembly of newly synthesized nucleocapsids and the packaging of RNA may occur in the cytoplasmic matrix and in the endoplasmic reticulum. Third, the new nucleocapsids may acquire envelopes and mature not only on the plasma membrane but also on the vacuole membranes inside the cell. Fourth, M1 virus may be released in 2 ways: nucleocapsids budding through the plasma membrane and mature virus-like particles moving through vacuole transportation (in CPV-0). Furthermore, HepG2 cells infected with M1 virus showed characteristics of apoptosis in the late stage, including swelling of the ER, condensation and marginalization of nuclear chromatin, and mitochondrial damage. In fact, we have also conducted similar observations on various cell lines, including Vero cells and other different kinds of cancer cells, suggesting that our observation of M1 life cycle is cell type independent (Supplementary Fig. S4). Notably, we also found many mature viral particles arrange in the form of lattice on the outside of the Vero cell membrane, which was not found in the HepG2 cells. It may due to the long infection time of Vero cell, or as virus-producing cells, infected Vero cell harvested more progeny virus.
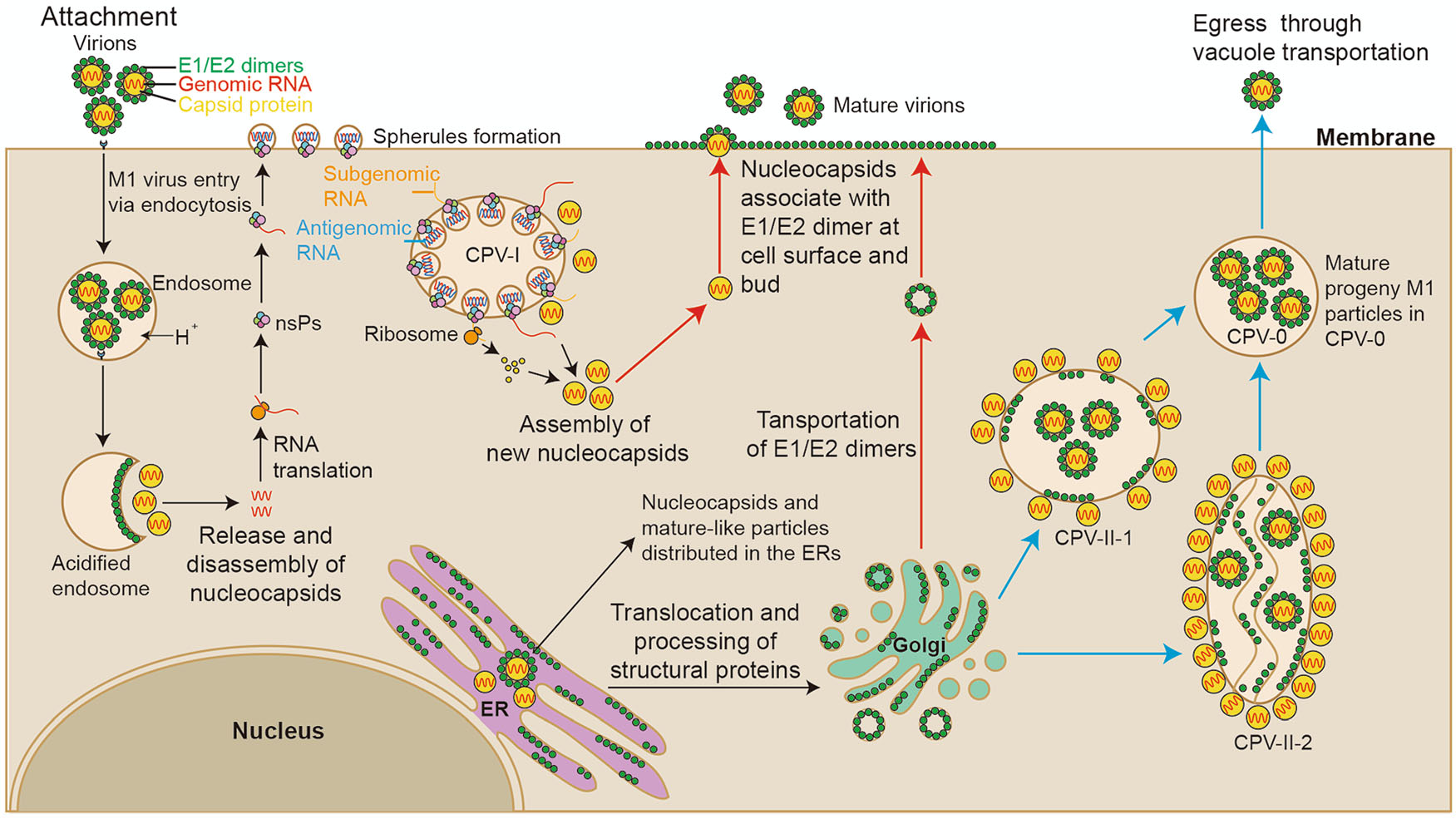
Figure 7. Schematic diagram of the M1 virus life cycle in HepG2 cells. According to our results and those of previous studies on alphaviruses, we have drawn a schematic diagram of the M1 virus life cycle. M1 viruses enter HepG2 cells via endocytosis and release their nucleocapsids in the cytoplasmic matrix for replication. Then, viral proteins and RNA are synthesized in CPV-I; capsid proteins and viral genomes are assembled into nucleocapsids in the ER and cytoplasmic matrix. There may be 3 ways that M1 virus matures: (1) envelope proteins are transported to the plasma membrane, and the M1 progeny mature through budding from the plasma membrane (red); (2) CPV-II-1 and CPV-II-2 form, envelope proteins are transported to the membrane of the CPV-IIs, nucleocapsids obtain envelopes through budding from the CPV-IIs (blue); and (3) nucleocapsids are assembled and the virus matures in the ER.
Ultrathin sectioning techniques have been widely applied to study subcellular structure, pathogen morphology and pathology for more than one-half century. The earliest reported alphavirus morphogenesis and pathology studies based on TEM were published in the 1960s (Acheson and Tamm 1967). Here, we applied the singleparticle analysis method to investigate the 2D structures and possible relationships among different kinds of particles during the M1 life cycle. With this technology, we have avoided some inaccuracies and thus obtained more accurate results. According to our observations, the precursors were scattered not only in the cytoplasmic matrix but also in ER cavities. Most literature on alphaviruses have reported that capsid proteins and viral genomes are assembled into nucleocapsids in the cytoplasmic matrix (Mendes and Kuhn 2018). Here, we also found nucleocapsids in the ER, suggesting that the ER may participate in nucleocapsid assembly in addition to the processing and transportation of viral envelope proteins. The 2D classification revealed that, in addition to nucleocapsid-like particles, another kind of particles, with an average diameter of 59 nm, were in ER; these particles were larger than the nucleocapsids and slightly smaller than the mature viruses. We suspect that these particles may also contain lipid bilayers and membrane proteins, but the structure of the envelopes may not have been completely processed.
Early studies suggested that CPV-I was derived from modified endosomes and lysosomes (Strauss et al. 1995). After viruses invade cells through the endocytic pathway, RNA replication and transcription occur on the membrane surface of the CPV-I (Spuul et al. 2010; Frolova et al. 2010). CPV-II was thought to be formed from the trans Golgi network (Soonsawad et al. 2010). Studies have shown that the tubular structures in CPV-II are composed of E1 and E2, the glycoproteins of the mature virus that form envelopes. The dimensions of the hexagonal array of glycoproteins within CPV-II are similar to the array of glycoproteins in the cell membrane, facilitating their budding and eventual virus assembly (Soonsawad et al. 2010). As a result, CPV-II likely serves as a transport vesicles and assembly sites for structural proteins. Our results showed two structures of CPV-IIs (CPV-II-1 and CPV-II-2) and both of them played key roles in M1 virus maturation and release. The 2D classifications of the particles inside CPV-II-1 and CPV-II-2 showed that M1 nucleocapsids may mature through budding from membranes of the CPV-II. Since viruses also need to be released outside the cell by budding, the question remains: how are M1 viruses that mature inside cells released out of cells? We found mature-like virus particles inside unique vacuoles, named CPV-0 and some of these vacuoles near the plasma membrane. According to these characteristics, we suspect that CPV-0 may be responsible for transport intracellular mature virus particles from CPV-II or ER out of the cells. More efforts are needed to prove the relationship between CPV-0 and CPV-II, and we also need to determine whether the M1 particles that mature inside cells are infectious.
In the late stage of infection, M1-infected cells gradually undergo apoptosis. Previous research indicated that M1 virus can trigger irresolvable ER stress and promote apoptosis (Lin et al. 2014). Here, we found that M1 virus infection also leads to swelling of the ER and apoptosis of the infected HepG2 cells. Although we observed apoptotic features, the plasma membrane remained intact, and no apoptotic body was found. Some studies have also shown that the formation of apoptotic bodies was negligibly observed in hepatocellular carcinoma tissues, and they considered that these apoptotic bodies were not essential for cell apoptosis. Instead, apoptotic cells were completely engulfed by neighboring tumor cells (Hang et al. 1996), which may be the reason that the body can rapidly and effectively eliminate dead cells without causing inflammatory reactions. The apoptotic cells infected by M1 virus were not engulfed by neighboring cells in our samples, which may be because most of the cells were in the later stage of M1 infection. Further efforts are needed to verify whether tumor cells infected by M1 virus are engulfed by uninfected neighboring tumor cells, thus causing further spread of M1 virus in a receptor-independent manner.
In conclusion, the life cycle of oncolytic M1 virus in hepatocellular carcinoma generally resembles that of other alphaviruses. Our data on the life cycle of M1 virus in tumor cells are expected to contribute to a greater in-depth understanding of oncolytic virus M1. Moreover, we now more deeply understand how M1 virus causes the death of tumor cells and the pathological features of M1-infected tumor cells, laying the foundation for us to further study M1 virus, modify the virus and use synergistic approaches to improve its oncolytic effect.
-
This research was supported by the Science and Technology Program of Guangzhou (201707020003), the Pioneering Talents Project of Guangzhou Development Zone, Guangdong Province (CY2018-012), the Guangzhou People's Livelihood Science and Technology Tackling Key Project (201803010113), the Young Teacher Training Program of Sun Yat-sen University (20ykpy22), and the National Natural Science Foundation of China (No. 81802536).
-
JD contributed to the experimental design and cellular experiments. JD and LN contributed to the EM observations, data analyses, manuscript writing and editing. XJ contributed to the 2D classification. JC, YL, JL, JH, GY and WZ contributed to the operations at the TEM facilities. YL and DC contributed to TEM sample preparation. CX, YL and CH contributed to the data analyses. JL and QZ contributed to the manuscript revisions. QZ contributed to the EM observations, data analyses, and project supervision.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.


















 DownLoad:
DownLoad: