HTML
-
Enterovirus 71 (EV71), a single-stranded RNA (ssRNA) virus belonging to the Picornaviridae family, serves as an important neurotropic virus responsible for hand-footmouth disease (HFMD) in infants or young children and can induce lethal neurological complaints (Ooi et al. 2010; Baggen et al. 2018). Since its first appearance in California in the USA in 1969 and association with epidemic HFMD in 1974 (Schmidt et al. 1974; Hagiwara et al. 1978), EV71 has recently become prevalent in many countries, especially those in the Asian-Pacific region (Ho et al. 1999; Xing et al. 2014; Solomon et al. 2010). To protect the public from EV71 contagion and to guide the development of anti-EV71 drugs, a deeper understanding of the mechanism of EV71 infection, especially how EV71 interacts with host cells, is needed.
The genome of EV71 is approximately 7.4 kb in length and contains an open reading frame (ORF) in its middle and untranslated regions (UTRs) at both its 3' and 5' ends (Hagiwara et al. 1984). After the virus infects host cells, the RNA genome is translated into a polyprotein that is subsequently divided into four structural proteins (VP1, VP2, VP3, VP4) and seven non-structural proteins (2A, 2B, 2C, 3A, 3B, 3C, 3D) (Baggen et al. 2018; Yuan et al. 2018).
As in other enteroviruses, the process of EV71 genome replication is well conserved and occurs on replication organelles (ROs). ROs may originate from Golgi membranes or the endoplasmic reticulum and are believed to protect viral RNAs from degradation by RNases or detection by cellular RNA sensors (Limpens et al. 2011; Suhy et al. 2000; van der Schaar et al. 2016). To form ROs for efficient viral genome synthesis, several non-structural viral proteins, including 2BC and 3A, may play a role in hijacking host factors, such as PI4KB, GBF1/Arf1, and ACBD3, to initiate RO assembly (Hsu et al. 2010; Dorobantu et al. 2014; Greninger et al. 2012; Arita 2014). The selective recruitment of host proteins regulates cellular lipid homeostasis, resulting in the rapid fabrication of a membrane-like structure, which serves as a raft for viral genome replication (van der Schaar et al. 2016; Belov and Kuppeveld 2012; Zhang et al. 2019). However, the elaborate components of ROs and their detailed assembly mechanism remain elusive (van der Schaar et al. 2016).
As the core enzyme of ROs, the EV71 3D protein, which functions as an RNA-dependent RNA polymerase (RdRp), is responsible for genome synthesis (Wu et al. 2010). During EV71 infection, 3D polymerase forms complex interactions with host factors, facilitating virus proliferation. 3D polymerase catalyses genome RNA elongation, which require assistance from host proteins including PCBP2, and hnRNPC (Bedard 2004; Blyn et al. 1996; Herold and Andino 2001; Brunner et al. 2010; Silvestri et al. 2006; Ertel et al. 2010). The activity of EV71 3D polymerase itself is also subject to regulation by host factors. Recent studies have shown that sumoylation can stabilize EV71 3D polymerase (Liu et al. 2016), and METTL3 can elevate the sumoylation level of 3D polymerase, thereby facilitating viral propagation (Hao et al. 2019).
To gain a deeper understanding of the mechanism of EV71 genome replication and based on the dependence of RO assembly and 3D polymerase function on complicated interactions with numerous host factors, we performed immunoprecipitation-mass spectrometry (IP-MS) and identified ANXA2 as a host protein that interacts with EV71 3D polymerase. ANXA2, a member of the annexin family that functions as a calcium-dependent membranebinding protein, is associated with immune regulation, tumour development, virus infection, etc. (Gerke and Moss, 2002; Hajjar 2015; Bećarević 2016; Schloer et al. 2018). ANXA2 has been verified to positively regulate several ssRNA and dsRNA viruses replication, including HCV, HPV, H5N1, and CMV (Backes et al. 2010; Saxena et al. 2012; Santos-Valencia et al. 2019; Ma et al. 2017; Derry et al. 2007; Woodham et al. 2014), and can also enhance EV71 infectivity in host cells (Yang et al. 2011). However, the interaction between ANXA2 and EV71 3D polymerase and how ANXA2 participates in EV71 genome replication have not yet been investigated. In this study, we verified that ANXA2 can interact with EV71 3D polymerase during EV71 infection. Subsequent experiments demonstrated that ANXA2 serves as a positive regulator of EV71 by promoting EV71 replication. Further experimentation showed that ANXA2 can interact with PI4KB and induce the production of phosphatidylinositol-4-phosphate (PI4P), which is critical for the assembly of EV71 ROs. Furthermore, 3D polymerase, ANXA2, and PI4KB form a higherorder protein complex located in ROs. ANXA2 can promote the interaction between 3D polymerase and PI4KB, which may be a method through which ANXA2 promotes the replication of EV71. Thus, ANXA2 is a key cellular protein that facilitates formation of the EV71 RNA replication complex.
-
RD (ATCC accession No. CCL-136), HEK293T (ATCC accession No. CRL-11268), and HeLa (ATCC accession No. CCL-2) cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Biological Industries) supplemented with 10% foetal bovine serum (Gibco, Grand Island, NY, USA) and 1% penicillin–streptomycin (Gibco). All cell lines were grown at 37 ℃ with 5% CO2 in a humidified environment.
The EV71 strain was provided by the State Key Laboratory of Virology (College of Life Sciences, Wuhan University). The virus was propagated in RD cells, and the titre was determined by plaque assay or 50% tissue culture infectious dose (TCID50) assay with RD cells.
-
The EV71 3D gene was amplified by PCR from the pACYC-EV71-FL plasmid, which contains the full-length EV71 cDNA clone and was a gift from Bo Zhang (Shang et al. 2013) (Wuhan Institution of Virology, CAS). The PCR products were subcloned into the EcoRI/BamHI sites of the p3XFlag-CMV-14 vector and SalI/NotI sites of the pCMV-HA vector. ANXA2 was amplified by PCR from a cDNA library derived from RD cells (the total cellular RNA was extracted with TRIzol [Invitrogen, CA, USA] and reverse transcribed with random or oligo dT primers plus M-MLV reverse transcriptase [Toyobo, Japan]). The PCR products were cloned into pCMV-HA at the SalI/NotI sites and pCDH-CMV-SF-IRES-Blast at the XhoI/BamHI sites. Truncated fragments of mutant ANXA2 (from amino acids 1–120, 1–192, 1–277, 55–357, 127–357, and 211–357) were obtained by PCR amplification of pCMVHA-ANXA2 and inserted into pCDH-CMV-SF-IRESBlast. The correct insertion of the sequence of interest into the expression plasmid was verified by DNA sequencing (Tiangen Biotech Co., Ltd., Wuhan, China). The primer sequences used in this study are shown in Supplementary Table S1.
-
In this study, the following primary antibodies were used: anti-ANXA2 rabbit polyclonal antibody (pAb, A12397, ABclonal, Wuhan, China), anti-EV71 3D mouse monoclonal antibody (mAb, GTX630193, GeneTex, CA, USA), anti-EV71 VP1 mAb (ab169442, Abcam, Cambridge, MA, USA), anti-dsRNA rJ2 mAb (MABE1134, Millipore, MA, USA), anti-PI4P mAb (Z-P004, Echelon Biosciences, UT, USA), anti-Flag M2 mAb (F1804, Sigma-Aldrich), antiHA mAb (H6908, Sigma-Aldrich), anti-a-Tubulin mAb (T6199, Sigma-Aldrich), and anti-GAPDH mAb (AC002, ABclonal). For immunoblotting, the secondary antibodies used were HRP-conjugated goat anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch Laboratories). Goat antimouse or anti-rabbit Alexa Fluor 488 and 555 (Thermo Fisher Scientific, MA, USA) and a DAPI staining solution (Beyotime, China) were used in the immunofluorescence assay.
-
The transfection of RD and HeLa cells was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. HEK293T cells were transfected with 0.1% (w/v) branched polyethyleneimine (PEI, SigmaAldrich). Briefly, HEK293T cells were seeded on plates one day prior to transfection and allowed to reach 60%–70% confluence at transfection. DNA (plasmids) and PEI were diluted in Opti-MEM (Gibco). After incubation at room temperature for 5 minutes (min), the diluted PEI was added to diluted DNA and incubated at room temperature for another 20 min. Then, the cell culture medium was replaced with FBS-free DMEM, and the PEI-DNA mixture was added to the cells. After incubation at 37 ℃ with 5% CO2 for 6 hours (h), the transfection mixture was replaced with complete DMEM (containing FBS), after which the cells were placed back into incubators.
EV71 was used to infect both RD and 293 T cells. In short, cells were incubated with the virus at 37 ℃ for 1 h. Next, the virus-containing medium was replaced with fresh medium, and the cells were returned to 37 ℃ incubators for the indicated duration. The viral infection titres varied among the different cell types and experiments.
-
HEK293T cells were transiently transfected with the p3XFlag-CMV-14-3D plasmid or empty vector. Thirty-six hours later, one group of cells was infected with EV71 (MOI = 10), while the other group of cells remained uninfected. Twelve hours post-infection, the cells were disrupted, and the lysates were incubated with anti-Flag M2 mAb (Sigma-Aldrich) for 2 h, followed by incubation with Protein G Agarose (Invitrogen) at 4 ℃ overnight. After washing with PBS three times, the agarose-bound samples were sent for mass spectrometry analysis on a TripleTOF@ 5600 system (performed by Procell Life Science & Technology Co., Ltd., Wuhan, China).
-
HEK293T cells were seeded in 10-cm cell culture dishes one day before lentiviral packaging. The following day, the HEK293T cells were co-transfected with the Δ8.9 packaging plasmid, vesicular stomatitis virus G protein expression plasmid (pVSVg) and ANXA2-overexpression plasmid (pCDH-CMV-ANXA2-SF-IRES-Blast) or empty control vector by using PEI. Two days later, supernatant containing the lentiviruses was harvested by centrifugation at 4 ℃ and 4000 rpm for 15 min and then filtered using a 0.45-lm filter. To construct stable cell lines, RD cells were seeded one day before infection in 6-well plates. Lentiviruses were added to the RD cells, and the plates were centrifuged at 37 ℃ and 2500 rpm for 2 h. The medium containing the lentiviruses was then replaced with fresh DMEM. Forty-eight hours later, blasticidin (25 μg/mL, Sigma-Aldrich) was added to screen for stable cell lines in which ANXA2 was overexpressed. Immunoblotting was finally performed to verify the stable expression of ANXA2 in the cell lines.
-
The target sequence used to generate a human ANXA2- knockout cell line by CRISPR-Cas9 was AAACTCGCCTACCAGAGAAGGACCC. First, the sequence was cloned into lenti-CRISPRv2, which was digested with BsmBI. To construct the knockout cell line, 12 μg of gRNA-expressing plasmid, 6 μg of the pVSVg plasmid and 6 μg of the Δ8.9 vector were co-transfected into 6 × 105 HEK293T cells. At 48 h post-transfection, the supernatant of the transfected cells, which contained the lentivirus, was collected for the following experiment. RD cells (or HEK293T cells) were plated into a 6-well culture plate. The next day, lentivirus was added to the cells. After 48 h, cells infected with the lentivirus were screened by using 1 μg/mL blasticidin. Four days later, single clones were screened by limiting dilution cloning. The knockout clones were verified by Western blotting.
-
HEK293T cells were seeded in 10-cm cell culture dishes one day before the experiment. The next day, plasmid for the expression of Flag/HA-tagged fusion protein or empty control vector was co-transfected into HEK293T cells with PEI. Forty-eight hours later, the cells were harvested and washed three times with PBS. Then, the cells were disrupted with cold RIPA lysis buffer (50 mmol/L Tris–HCl, pH 7.5, 150 mmol/L NaCl, 2 mmol/L EDTA, pH 8.0, 0.5% NP-40 and a protease inhibitor cocktail [Roche], 1 mmol/L PMSF) at 4 ℃ for 30 min, followed by centrifugation at 4 ℃ and 12, 000 rpm for 15 min. Subsequently, the supernatant of the cell lysates was mixed with anti-Flag M2 affinity gel (Sigma-Aldrich) and incubated at 4 ℃ overnight on a vertical rocker. The gel was then washed with cold RIPA lysis buffer three times and analysed by Western blotting with anti-Flag or anti-HA antibody.
For endogenous co-IP experiments, RD cells were seeded in 10-cm cell culture dishes one day prior to the experiment and infected with EV71 (MOI = 0.5) for 12 h. The cells were harvested, washed with PBS three times and disrupted with RIPA lysis buffer. Next, the cell lysates were incubated with anti-EV71 3D antibody or control goat IgG (ABclonal) for 2 h. After that, Protein G Agarose (Invitrogen) was added and incubated at 4 ℃ overnight. The agarose was then washed and analysed by Western blotting with anti-EV71 3D or anti-ANXA2 antibody.
-
Briefly, cell lysates or co-IP products were separated by SDS-PAGE and then transferred to a nitrocellulose membrane (Pall). The membrane was blocked with 5% skim milk (BD Biosciences, San Jose, CA, USA) dissolved in TBST (150 mmol/L NaCl, 20 mmol/L Tris base, 0.05% Tween 20), followed by incubation with primary antibodies at 4 ℃ overnight. After washing three times with TBST, the membrane was incubated with HRP-conjugated secondary antibodies at room temperature for 1 h. The protein signals were detected by using ECL reagents (Millipore) on a GE Amersham Imager 600 (Milwaukee, WI, USA).
-
HeLa cells were seeded on coverslips in 24-well plates one day before the experiments. The next day, the cells were co-transfected with pCMV-HA-ANXA2 and p3XFlagCMV-14-3D or the empty control vector by using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, the cells were fixed with 4% paraformaldehyde for 15 min at room temperature and washed with PBS three times. Then, the cells were permeabilized with 50 mg/mL digitonin dissolved in PBS for 5 min, washed with PBS three times, blocked with 5% BSA in PBS for 30 min, and incubated with anti-Flag mouse mAb (1:200) and anti-HA rabbit pAb (1:200) at 4 ℃ overnight. After washing five times with PBS, the cells were stained with anti-mouse Alexa Fluor 488 and anti-rabbit Alexa Fluor 552 at room temperature for 1 h. A diamidino-2-phenylindole (DAPI) solution was then added and incubated for 5 min to counterstain the nuclei. The coverslips were washed with PBS five times, fixed onto microslides and finally analysed using a Leica SP8 lightning confocal microscope.
To assess the endogenous co-localization of ANXA2 with EV71-3D, PI4P or dsRNA, RD cells were infected with EV71 (MOI = 1) for the indicated durations. The cells were fixed and stained with anti-ANXA2 rabbit pAb (1:50), anti-EV71 3D mouse mAb (1:100), anti-PI4P mouse mAb (1:100) or mouse anti-dsRNA J2 mAb (1:200).
-
Total cellular RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Three micrograms of RNA was reverse transcribed into cDNA using Toyobo RT kits with a genomic DNA remover (A1172K). After that, qPCR was performed in a reaction volume of 10 μL containing 5 μL of SYBR Green master mix (Toyobo), 1 μL of the mixed primers, and 4 μL of diluted cDNA samples and carried out on a QuantStudioTM 6 Flex system. Relative qPCR data were analysed by the double delta Ct method with the housekeeping gene GAPDH.
For absolute qPCR to determine the EV71 viral particle titre in each supernatant, viral nucleic acid isolation kits (Simgen, China) were used to extract the EV71 genome. Serial dilutions of the plasmid pACYC-EV71-FL served as standard samples from which a standard curve was plotted. The EV71 copy number in the supernatant was then calculated based on the standard curve. All primer sequences used in the experiments are shown in Supplementary Table S1.
-
Statistical analyses of the qRT-PCR data were performed using Microsoft Excel software. Graphs represent the means and standard deviations. The two-tailed Student's t-test was used for two-group comparisons. P values < 0.05, < 0.01, and < 0.001 were used to indicate significance.
Cell Lines and Viruses
Plasmids and Constructs
Antibodies and Reagents
Transfection and Infection
Proteome Analysis
Establishment of a Stable ANXA2-Expressing RD Cell Line
Generation of a CRISPR-Cas9 Knockout Cell Line
Co-immunoprecipitation (co-IP)
Immunoblotting
Immunofluorescence
RNA Extraction and Quantitative Real-Time PCR (RT-qPCR)
Statistical Analyses
-
To better understand the mechanism of EV71 replication, we performed an IP-MS experiment to explore the host proteins that interact with EV71 3D polymerase. The recombinant 3D-Flag protein was overexpressed in HEK293T cells, and proteins that interacted with recombinant 3D-Flag were enriched by immunoprecipitation and analysed by LC–MS/MS either with or without EV71 infection (Fig. 1A). Different candidate protein interactors from the two groups were compared and visualized with a Venn diagram. From the overlap, we identified ANXA2 as a host protein that may interact with EV71 3D polymerase (Fig. 1B).
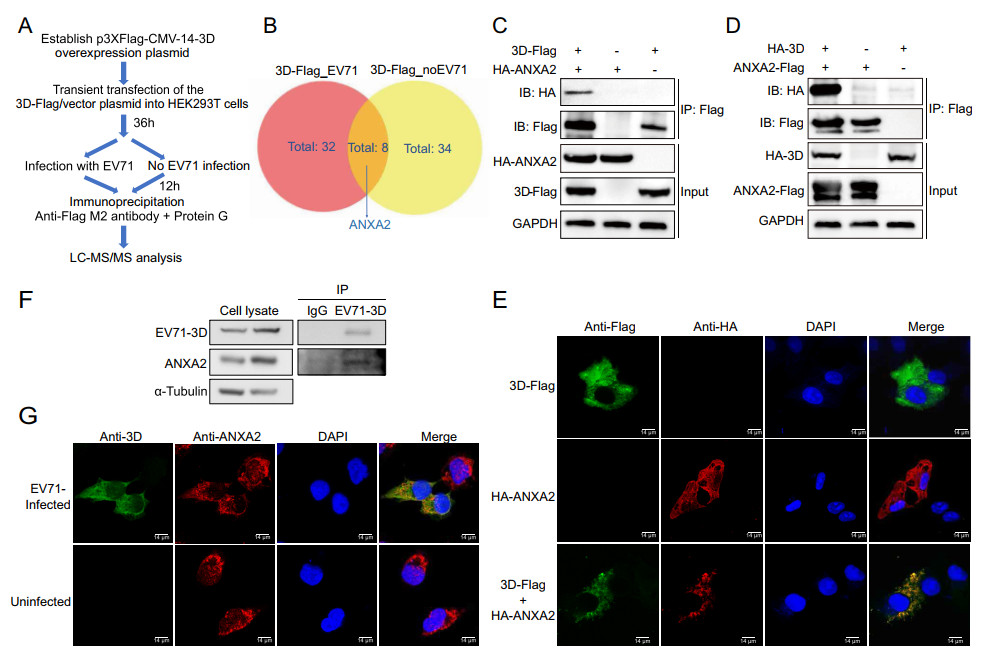
Figure 1. Identification of ANXA2 as a host protein interacts with EV71 3D polymerase. A Workflow for IP-MS screening of host proteins interacted with EV71 3D polymerase. B Venn diagram shows the differential interactor candidates for 3D-Flag fusion protein with or without the context of EV71 infection. ANXA2 was identified from the overlaps. 3D-Flag_EV71, overexpression of 3D-Flag fusion protein in HEK293T cells and infecting with EV71. 3D-Flag_no EV71, overexpressing 3D-Flag fusion protein without virus infection. C HEK293T cells were transfected with plasmids expressing either 3D-Flag or HA-ANXA2 or co-transfected with both of the plasmids. Cell lysates were immunoprecipitated with anti-Flag M2 affinity gel and indicated proteins were analyzed by Western blot. Similar coimmunoprecipitation was performed in (D) with plasmids switched to HA-3D and ANXA2-Flag. E Exogenous colocalization was identified in Hela cells with red represents HA-ANXA2 and green represents 3D-Flag. DAPI was used to show nuclei (blue). Images were visualized under confocal microscopy. Scale bar = 14 μm. F Coimmunoprecipitation was performed in RD cells after 12 h EV71 infection (MOI = 0.5) by using endogenous anti-3D antibody. IgG was used as control. Indicated proteins were detected by Western blot. G Endogenous colocalization between ANXA2 and EV71 3D polymerase was detected in RD cells after 12 h infection of EV71 (MOI = 0.5) by using anti-3D and anti-ANXA2 primary antibody. Uninfected cells served as control. Green fluorescence stained 3D polymerase and red fluorescence for ANXA2, Blue represents nuclei. Scale bar = 14 μm.
To confirm the interaction between ANXA2 and 3D polymerase, we first exogenously expressed the 3D-Flag and HA-ANXA2 fusion proteins in HEK293T cells and performed a co-immunoprecipitation (co-IP) assay to capture them in complex. As shown in Fig. 1C, HA-ANXA2 specifically co-immunoprecipitated with 3D-Flag, but no nonspecific binding was observed for the control groups (with only 3D-Flag or HA-AXNA2). To exclude the potential influence of the affinity tags on the protein–protein interaction, we switched the tags of the two proteins and repeated the co-IP assay. Consistent with the above finding, after switching the two protein tags, HA-3D could still be co-immunoprecipitated with ANXA2-Flag (Fig. 1D). To further investigate whether ANXA2 and 3D polymerase are located in the same cellular compartment, we performed a co-immunofluorescence assay in HeLa cells. The results showed that 3D-Flag and HA-ANXA2 co-localize in the same cytoplasmic region in the HeLa cells (Fig. 1E). These results demonstrated the strong interaction between exogenous ANXA2 and 3D polymerase.
We next set out to probe whether endogenous ANXA2 can interact with 3D polymerase during the infection of RD cells with EV71. After treatment with EV71 (MOI = 0.5) for 12 h, an anti-EV71 3D antibody was used to enrich EV71 3D polymerase. Unlike the results in the goat IgG control group, endogenous ANXA2 co-immunoprecipitated with 3D polymerase (Fig. 1F). A parallel immunofluorescence assay indicated the co-localization of endogenous ANXA2 with 3D polymerase in the same region during EV71 infection (Fig. 1G). Taken together, the above observations suggest that ANXA2 serves as a novel host factor that interacts with viral 3D polymerase during EV71 infection.
-
To examine the impact of ANXA2 on EV71 replication, we first constructed an RD cell line that stably expressed ANXA2 by employing lentivirus infection. Western blot analysis verified that the recombinant ANXA2-Flag protein was steadily expressed in the stable RD cells (Fig. 2A). We then sought to compare EV71 infection efficiencies in the stable ANXA2-overexpressing RD cell line and wild-type RD cell line. Both cell lines were treated with EV71 at an equal MOI at 0 h. Within 36 h post-infection (hpi), RTqPCR showed an increase in the ANXA2 mRNA level in the stable ANXA2-overexpressing RD cell line comparing with the wild-type RD cell line, which further confirmed that ANXA2 was continuously overexpressed (Fig. 2B). To evaluate EV71 infection efficiency, we probed viral protein expression levels, relative viral RNA levels, and viral particle titers in the supernatant. We observed elevated expression of the EV71 VP1 capsid protein and an increase in the relative EV71 RNA level at 12, 24, and 36 hpi in the stable RD cell line (Fig. 2C, 2D). Accordingly, the mature viral particle titers in the supernatant at 24 and 36 hpi was significantly higher for the ANXA2-overexpressing RD cell line than in the wild-type RD cell line (Fig. 2E). This evidence indicated that the overexpression of ANXA2 in RD cells facilitated EV71 genome replication, promoted viral protein expression, and resulted in more mature viral particles being released into the extracellular space.
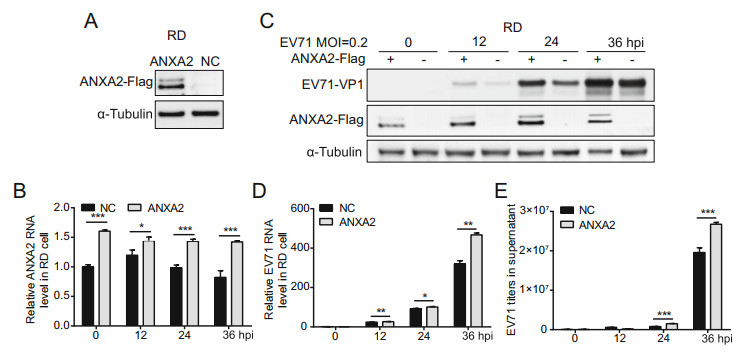
Figure 2. Overexpression of ANXA2 promotes EV71 replication. A Establishment of ANXA2-Flag stable overexpression cell lines was verified by Western-blot. NC, negative control. B Relative ANXA2 RNA levels in cells for each time points were quantified by RT-qPCR and normalized to endogenous GAPDH RNA level. C RD cells were infected with EV71 (MOI = 0.2). At 0, 12, 24, 36 h post infection (hpi), EV71 VP1 and ANXA2-Flag protein expression levels in cells were detected by western-blot by using anti-VP1, anti-Flag and anti-α-Tubulin antibodies. D Relative EV71 RNA levels in cells for each time points were quantified by RT-qPCR and normalized to endogenous GAPDH RNA level. E Titer of EV71 viral particles in supernatant at indicated time points were determined via RT-qPCR and calculated according to standard curve. B, D, E Data were shown with mean ± s.d., n = 3, student's t-test, *P < 0.05, **P < 0.01, ***P < 0.001.
As the overexpression of ANXA2 upregulated EV71 infection, we hypothesized that ANXA2 knockout would have the opposite effect. To verify this hypothesis, we generated an ANXA2-KO RD cell line by clustered regularly interspaced short palindromic repeat (CRISPR)-mediated disruption. As illustrated in Fig. 3A, no ANXA2 protein was detected in the ANXA2-KO RD cell line. Moreover, the mRNA level of ANXA2 was significantly lower than that in the control group, indicating successful construction of the ANXA2-KO RD cell line (Fig. 3B). In agreement with our hypothesis, depletion of ANXA2 reduced the protein expression of EV71 VP1 (Fig. 3C) and inhibited EV71 genome synthesis (Fig. 3D). We also observed a significant decrease in EV71 viral particles release in the supernatant for the ANXA2-KO RD cell line (Fig. 3E). In a virus synchronization experiment (Fig. 3F), we observed no significant difference in relative EV71 RNA level between the control RD cell line and ANXA2- KO RD cell line in the first 6 h after infection. In contrast, synthesis of the EV71 RNA genome was significantly reduced in the ANXA2-KO RD cell line 6 h post-infection, which further confirmed that ANXA2 acts during the genome replication phase of the EV71 life cycle. In general, ANXA2 deletion significantly inhibited EV71 replication in RD cells, indicating that ANXA2 is an important positive regulator of the replication of EV71.
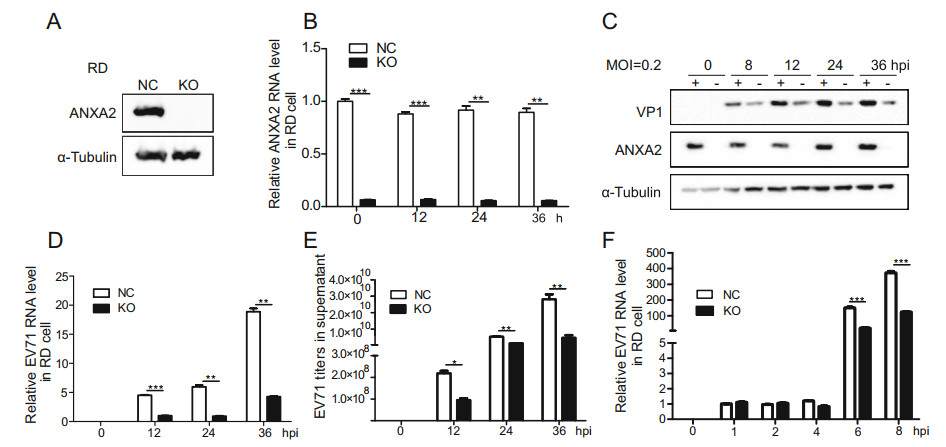
Figure 3. Knockout of endogenous ANXA2 inhibits EV71 proliferation. A Knockout efficiency of ANXA2 in RD cells. B Relative ANXA2 RNA levels in cells for each time points were quantified by RT-qPCR and normalized to endogenous GAPDH RNA level. C ANXA2-KO RD cells were infected with EV71 (MOI = 0.2). At 0, 8, 12, 24, 36 h post infection (hpi), EV71 VP1 and ANXA2 protein expression levels in cells were detected by western-blot by using anti-VP1, antiANXA2 and anti-α-Tubulin antibodies. D Relative EV71 RNA levels in cells at 0, 12, 24, 36 hpi were detected by RT-qPCR and normalized to GAPDH RNA levels. E EV71 viral titers in supernatant were quantified via RT-qPCR and calculated based on standard curve. F RD cells were infected with EV71 at an MOI of 1 PFU/cell, The treatment time was 4 ℃ for 30 min and then infected at 37 ℃ for 1 h. Relative EV71 RNA levels in cells at 0, 1, 2, 4, 6, 8 hpi were detected by RT-qPCR and normalized to GAPDH RNA levels. (C–E) Data were shown with mean ± s.d., n = 3, student's t-test, *P < 0.05, **P < 0.01, ***P < 0.001.
-
Similar to other members of the annexin family, ANXA2 is composed of a highly variant N-terminal domain (NTD) and conserved C-terminal core domain consisting of four annexin repeats (ANX repeats) (Mirsaeidi et al. 2016; Bharadwaj et al. 2013). To determine the domain of ANXA2 that interacts with 3D polymerase, we generated a series of truncated ANXA2 constructs (amino acids 1–120, 1–192, 1–277, 55–357, 127–357, and 211–357) based on the NCBI reference sequence NP_001002858.1 (Fig. 4A). All of the truncated constructs were linked to the Flag tag and co-transfected with HA-3D into HEK293T cells, with full-length ANXA2 and empty vector serving as controls. Surprisingly, a co-IP experiment revealed that HA-3D could be co-immunoprecipitated by all six truncated constructs, suggesting that the annexin domain, contained in all of the constructs, is responsible for the binding of ANXA2 to 3D polymerase, while the N-terminal domain is not necessary for this interaction to occur (Fig. 4B).

Figure 4. Annexin domain of ANXA2 mediates the interaction with 3D polymerase and the promotion of EV71 replication. A Schematic designing of ANXA2 truncated mutants. B Mapping the interacted domain of ANXA2 to 3D polymerase. All of the six truncated mutants (1–120, 1–192, 1–277, 55–357, 127–357, 211–357) were linked with Flag tag and corresponding plasmids, along with plasmids expressing full-length ANXA2 and empty vectors, were co-transfected with HA- 3D overexpression plasmid in HEK293T cells, respectively. Cell lysates were harvested after 48 h transfection and co-immunoprecipitated with anti-Flag M2 affinity gel. Indicated proteins were detected by western-blot. *, distinguishing bands for truncated mutants from visible non-specific bands. C Truncated mutants which excluded NTD domain (55–357, 127–357, 211–357) plus 1–277 were transfected and expressed in HEK293T cells. 24 h later, EV71 was treated with MOI = 0.5. EV71 VP1 and truncated mutants were then detected by western-blot at 24 hpi.
This phenomenon gave rise to our hypothesis that the annexin domain plays an essential role in the ANXA2- mediated promotion of EV71 replication. To test this hypothesis, truncated constructs excluding the N-terminal domain (amino acids 55–357, 127–357, and 211–357) were expressed in HEK293T cells that were then infected with EV71 (MOI = 0.5). At 24 h post-infection, the EV71 VP1 capsid protein expression level was measured. As shown in Fig. 4C, all three truncated constructs induced the upregulation of EV71 VP1 expression. Interestingly, with shortening of the annexin domain from four repeats (amino acids 55–357) to two repeats (amino acids 211–357), the expression level of VP1 gradually increased (Fig. 4C, lanes 4–6). Moreover, a previous study reported that the fourth annexin repeat of ANXA2 can interact with EV71 VP1 and facilitate the attachment of EV71 to host cells (Yang et al. 2011). However, our additional test for the truncated construct from amino acids 1–277 indicated that the absence of the fourth annexin domain did not diminish the ability of ANXA2 to enhance EV71 replication (Fig. 4C, lane 3).
-
RNA viruses induce the formation of special membrane structures for genome replication. These structures are often referred to as ROs. ROs exhibit lipid compositions distinct from those of other cellular membranes. In many picornaviruses, phosphatidylinositol-4-phosphate (PI4P) is a marker of ROs (Banerjee et al. 2018).
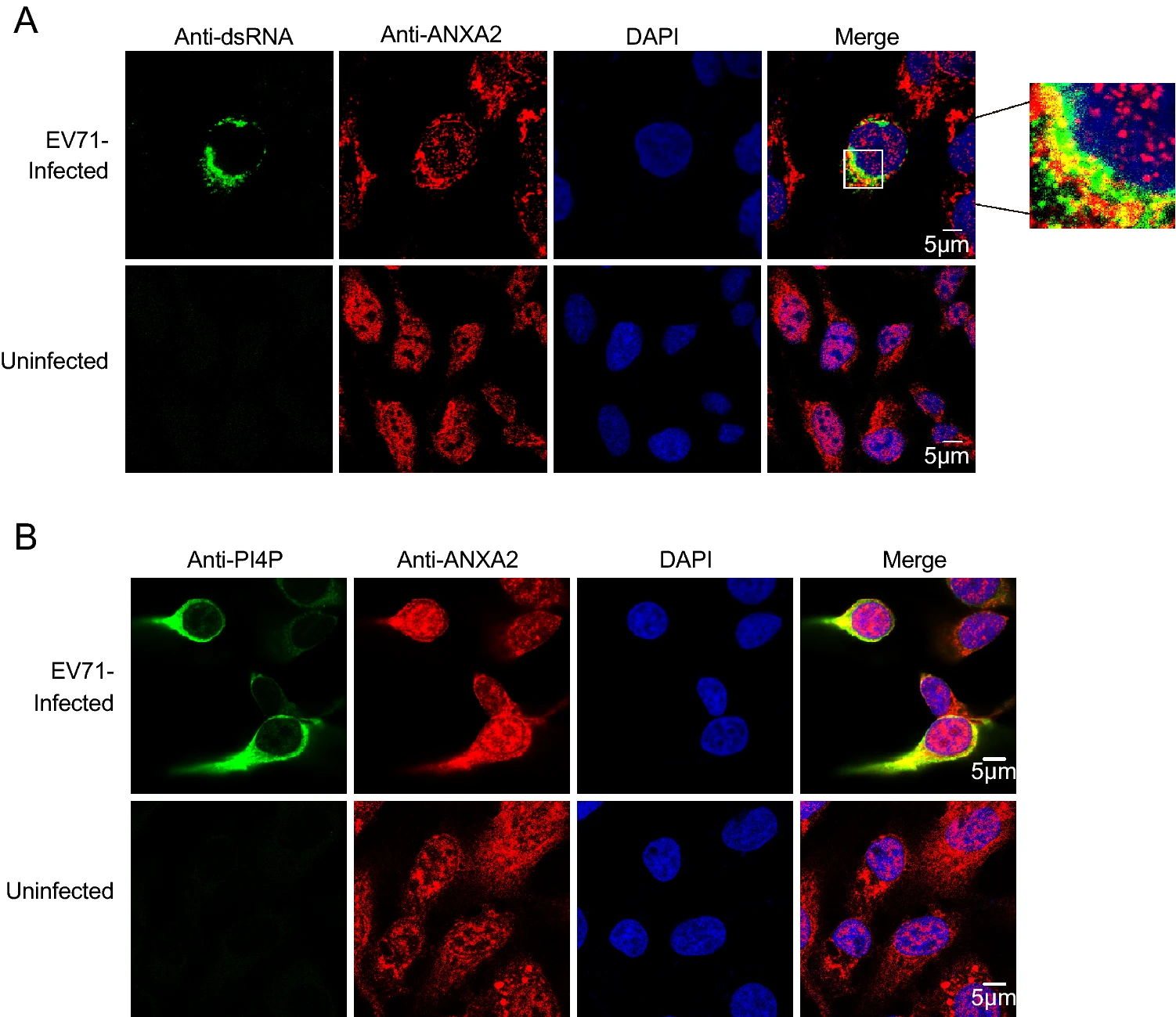
A previous study noted that the annexin domain is known for its membrane-binding capacity (Gerke and Moss 2002). As we observed that the annexin domain itself can facilitate EV71 replication, we speculated that ANXA2 may also be involved in EV71 ROs via the annexin domain. Double-stranded RNA (dsRNA) is an intermediate product produced during duplication of the ssRNA genome (Jacobs and Langland 1996). The synthesis of dsRNA occurs on ROs (Jacobs and Langland 1996). To verify whether ANXA2 is a part of EV71 ROs, we harvested EV71-infected RD cells at 6 h post-infection and performed a co-immunofluorescence assay to determine whether ANXA2 would co-localize with dsRNA. As shown in Fig. 5A, ANXA2 and dsRNA appeared in the same region of the cytosolic compartment, which indicates that ANXA2 participates in EV71 ROs.
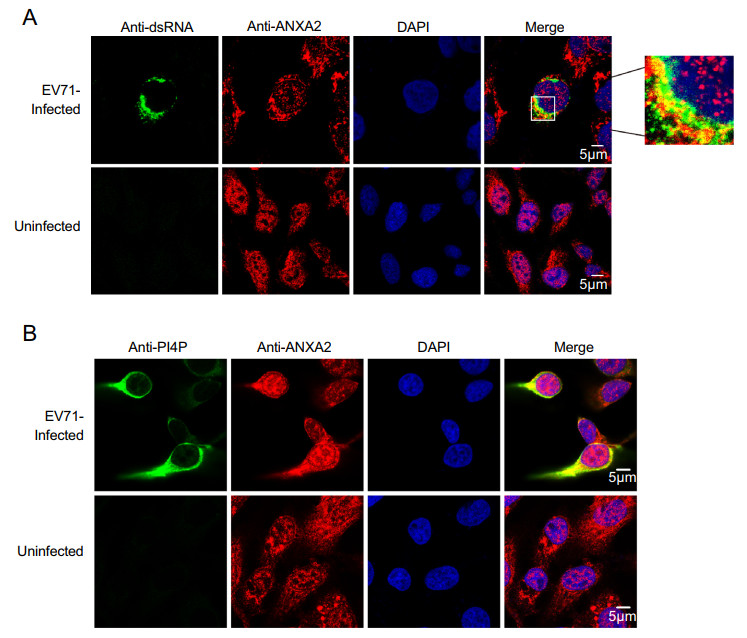
Figure 5. ANXA2 located to the replication organelles of EV71. A RD cells were infected with EV71 at an MOI of 1 PFU/cell. After 6 h, the cells were fixed and labeled with anti-dsRNA (green) and anti-ANXA2 (red) antibodies. The cells were labeled with anti-PI4P (green) (B) and anti-ANXA2 (red) antibodies.
Moreover, phosphatidylinositol 4-phosphate (PI4P), the enrichment of which in ROs provides a lipid microenvironment (Hsu et al. 2010), also showed clear co-localization with ANXA2 at 6 h post-infection with EV71 (Fig. 5B). These results suggest that ANXA2 indeed participates in EV71 ROs formation.
-
Previous studies have reported that EV71 replication is dependent on phosphatidylinositol 4-kinase Ⅲ (PI4KB). In general, PI4KB is recruited to replication sites by directly interacting with viral proteins or with the help of cytokines such as GBF1/ARF1 or ACBD3. Then, PI4KB yields phosphatidylinositol-4-phosphate (PI4P) on ROs, after which PI4P binds and induces conformational changes in the RdRp to modulate RdRp activity (Li et al. 2019). Therefore, we asked whether ANXA2 can interact with PI4KB, which produces PI4P. We exogenously expressed the HA-ANXA2 and Flag-PI4KB fusion proteins in HEK293T cells and performed a co-IP assay as well as an immunofluorescence assay. As illustrated in Fig. 6A–6B, HA-ANXA2 specifically co-immunoprecipitated and colocalized with Flag-PI4KB. These results showed an exogenous interaction between ANXA2 and PI4KB. Subsequently, we investigated the effect of ANXA2 overexpression on PI4P. As shown in Fig. 6C, overexpression of ANXA2 in RD cells upregulated PI4P. A further experiment showed that PI4P was significantly decreased in EV71-infected ANXA2-KO RD cells (Fig. 6D).
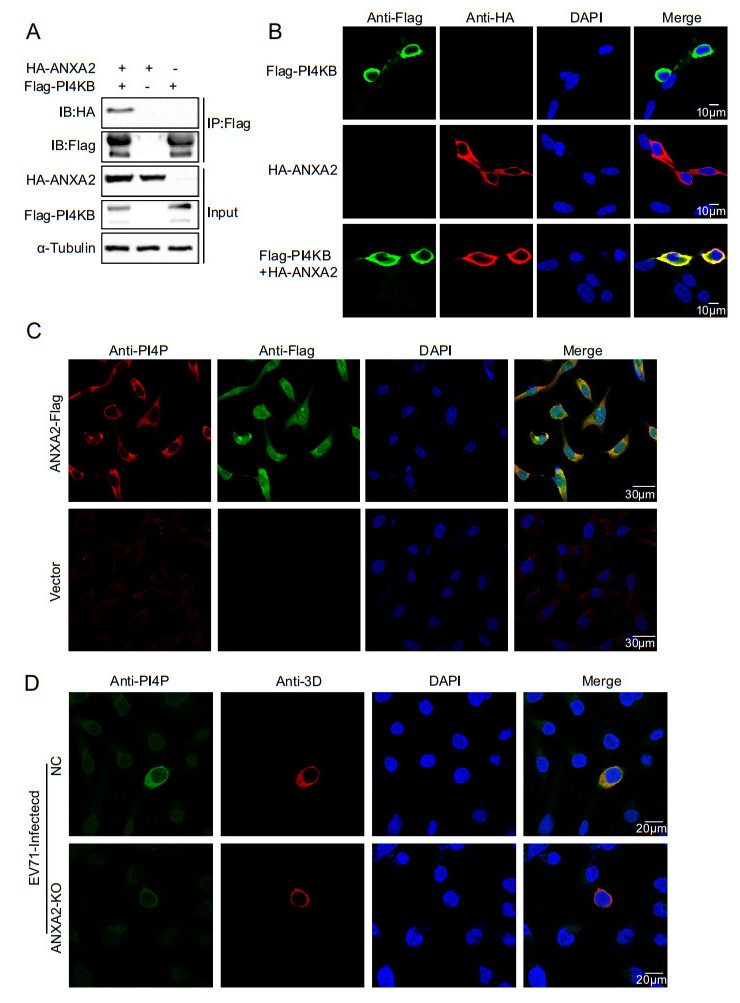
Figure 6. ANXA2 interacts with PI4KB and promotes the formation of PI4P. A HEK293T cells were transfected with plasmids expressing either FlagPI4KB or HA-ANXA2 or cotransfected with both of the plasmids. Cell lysates were immunoprecipitated with antiFlag M2 affinity gel and indicated proteins were analyzed by Western blot. B Exogenous colocalization was identified in HEK293T cells with red represents HA-ANXA2 and green represents FlagPI4KB. DAPI was used to show nuclei (blue). C The stable ANXA2-overexpressing RD cell lines and control cells were analyzed for formation of endogenous PI4P. D ANXA2 KO RD cells and control cells were infected with EV71 at an MOI of 1 PFU/cell. At 6 h post infection, the cells were fixed and labeled with antibodies against PI4P (green) and 3D (red).
-
Since ANXA2 interacts with 3D polymerase and PI4KB and all of these proteins were shown to be located at the replication sites of EV71, the three proteins likely to form a large protein complex on ROs. Thus, we further verified whether 3D polymerase and PI4KB interact. We exogenously expressed the HA-3D and Flag-PI4KB fusion proteins in HEK293T cells and performed a co-IP assay as well as an immunofluorescence assay. Interestingly, as shown in Fig. 7A–7B, HA-3D alsospecifically co-immunoprecipitated and co-localized with Flag-PI4KB. These results strongly suggest that 3D polymerase, ANXA2, and PI4KB form a higher-order protein complex located in ROs. To clarify the role of ANXA2 in this large protein complex, an ANXA2-KO HEK293T cell line (Fig. 7C) was transfected with Myc-ANXA2, Flag-PI4KB, and HA-3D expression plasmids. After 36 h, the cell lysates were immunoprecipitated with antibody against Flag tag. As shown in Fig. 7D, although 3D and ANXA2 were expressed at comparable levels, 3D alone weakly bound PI4KB (lane 2). Co-expression of ANXA2 led to an increase in the binding of 3D to PI4KB (lane 1), suggesting that ANXA2 facilitates the interaction of 3D polymerase and PI4KB in the absence of other viral proteins.
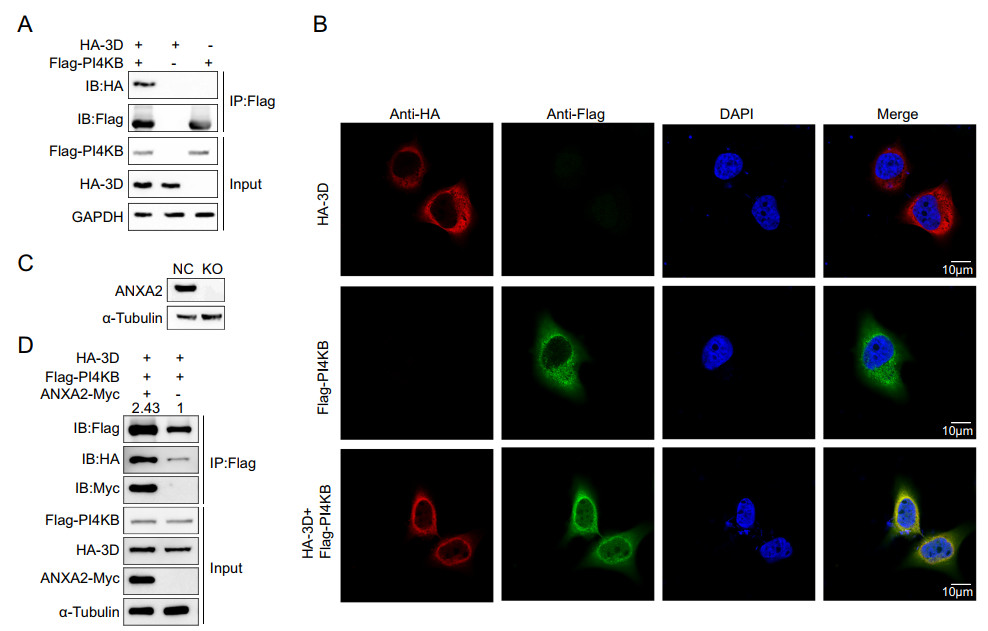
Figure 7. EV71 3D, ANXA2, and PI4KB form a protein complex. A HEK293T cells were transfected with plasmids expressing either Flag-PI4KB or HA-3D or co-transfected with both of the plasmids. Cell lysates were immunoprecipitated with anti-Flag M2 affinity gel and indicated proteins were analyzed by Western blotting. B Exogenous colocalization was identified in Hela cells with red represents HA-3D and green represents Flag-PI4KB. DAPI was used to show nuclei (blue). Images were visualized under confocal microscopy. C Knockout efficiency of ANXA2 in HEK293T cells. D ANXA2 KO HEK293T cells were transfected with plasmids expressing FlagPI4KB, HA-3D and ANXA2-Myc as indicated. After 36 h, the cell lysates were immunoprecipitated (IP) using anti-Flag, followed by Western blotting using anti-Flag antibodies. IB, immunoblotting.
Identification of ANXA2 as a Host Protein that Interacts with EV71 3D Polymerase
ANXA2 Serves as Positive Regulator of EV71 Replication
The Annexin Domain of ANXA2 Mediates the Interaction of ANXA2 with 3D Polymerase and Promotes EV71 Replication
ANXA2 Participates in EV71 ROs Formation
ANXA2 Interacts with PI4KB and Promotes the Formation of PI4P
ANXA2 Forms a Protein Complex with 3D Polymerase and PI4KB, which Then Stimulates the PI4KB-3D Polymerase Interaction
-
ANXA2 is known to be involved in many stages of virus infection, including virus entry, replication, and assembly (Aliyu et al. 2019). In this study, we found that EV71 3D polymerase interacts with ANXA2 and promotes EV71 replication. First, we verified the interaction between EV71 3D polymerase and ANXA2 by an immunoprecipitation experiment. Moreover, both exogenous and endogenous ANXA2 and EV71 3D polymerase could co-localize in the cytoplasm. We subsequently verified that ANXA2 can positively regulate EV71 infection. Overexpression of ANXA2 in different cell lines promoted EV71 VP1 protein expression, elevated EV71 RNA levels and increased the number of viral particles in the supernatant. Correspondingly, ANXA2 knockout inhibited the replication of EV71, which further confirmed our finding. Further experiments showed that the annexin domain of ANXA2 mediates the interaction with 3D polymerase and the ability of ANXA2 to promote EV71 replication.
PI4KB is a key enzyme in enterovirus replication. Previous studies have shown that enterovirus 3A recruits PI4KB into viral ROs under the action of the host factor acyl-coenzyme A binding domain containing 3 (ACBD3) (Greninger et al. 2012; Xiao et al. 2017). PI4KB produces PI4P, which forms a lipid microenvironment at ROs, regulates the activity of the RdRp, and promotes virus replication. In addition, changes in the curvature of the RO membrane protect the virus from host defences (Belov and Kuppeveld 2012). In our study, the host protein ANXA2 was found to interact with PI4KB and localize in EV71 ROs. We also noticed that the overexpression and knockout of ANXA2 upregulated and decreased PI4P levels, respectively, and that this regulatory process was virus independent. However, this regulatory process may be exploited by EV71, which may be a method by which ANXA2 promotes EV71 replication. Since PI4KB plays an important role in the ROs of enteroviruses and even positive-strand RNA viruses, whether ANXA2 and PI4KB interact in other viruses is worthy of further exploration.
We also dissected the potential mechanism by which the interaction of ANXA2 and 3D polymerase facilitates EV71 replication. We observed that 3D polymerase, ANXA2, and PI4KB are located in ROs and form a protein complex. 3D polymerase could interact with PI4KB in the absence of ANXA2, indicating that they may directly interact or that other host factors mediate their interaction. Interestingly, ANXA2 could interact with both PI4KB and 3D polymerase in this protein complex and enhance the interaction between 3D polymerase and PI4KB, suggesting that ANXA2 may act as a scaffold protein on ROs that helps anchor the key enzymes in the process of EV71 replication to the membrane to form the viral replication complex. Thus, ANXA2 is a key cellular protein that assists in the assembly of EV71 ROs.
In summary, enteroviruses reshape cell membranes to form replicating organelles under the action of viral proteins and host factors. This study highlights the essential role of ANXA2 in the replication of EV71 and may yield therapeutic strategies for enterovirus-induced diseases.
-
We thank Dr. Bo Zhang (Wuhan Institution of Virology, CAS) for generously providing pACYC-EV71-FL plasmid; Dr. Mingzhou Chen (Wuhan University, China) for pCAGGS-FlagPI4KB plasmid. This study was supported by the National Science Foundation of China (81971976, 81772236), Major Project of Technology Innovation Program of Hubei Province (2018ACA123).
-
QZ, SL, SW and KL designed the study. QZ and SL performed the research, analysed data, and wrote the paper. SW and KL checked and finalized the manuscript. All authors discussed the results and revised the manuscript.
-
The authors declare that they have no conflicts of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.















 DownLoad:
DownLoad: