-
Rice gall dwarf disease is one of the most serious diseases in rice in Southern China, Thailand, Korea and other Southeastern Asian countries (2, 11, 13). It was first discovered in Thailand in 1979 (11). It has erupted several times in Southern China since 1980s, and is still represents one of the main rice diseases in these regions. The symptoms include stunting of the plants, gall formation along leaf blades and sheaths, and dark green discoloration (2, 11).
Rice gall dwarf virus (RGDV), the pathogen of this disease, is grouped in the genus Phytoreovirus (11, 12) in the family of Roeviridae (12). The virus multiplies in leafhopper vectors such as Nephotettix nigropictus and N. cinticeps. It is transmitted in a persistent manner and restricted within the phloem cells. RGDV, as well as Rice dwarf virus (RDV) and Wound tumor virus (WTV), is an icosahedral double-shelled particle approximately 65-70 nm in diameter that contains 12 genome segments of dsRNA (4, 10, 12), which have been named S1 to S12 according to their electrophoretic mobility in polyacrylamide gels.
RNAs of all 12 genome segments of RDV and WTV have been sequenced (8, 9, 15). But in the case of RGDV, only the nucleotide sequences of genome segments S2 (8), S3 (15), S5 (3), S8 (10), S9 (4), S10 (10), and S11 (9) of Thailand isolate have been reported.
In China, this disease broke out first in Xinyin County, Guangdong Province. Fortunately, it was successfully controlled by controlling its principal vectors at the early stage of rice growth. But in recent years, the presence of RGDV has also been discovered in other Counties, such as GaoZhou and DeQin, in Guangdong Province.
To determine the epidemiology of the rice gall dwarf disease, an investigation was carried out in China. This disease was found to be present in other two counties of Guangdong Province, BoLuo and CongHua, which revealed that RGDV had been continuing to spread. How RGDV has been introduced to these areas remains unknown since the leafhopper could not migrate for long distances and raised the additional question as to why is RGDV sporadic in Thailand while epidemic in South China? Previous reports showed that some differences were found between isolates from China and from Thailand in their principal vectors and artificial inoculation hosts (2). To understand the relationships between Chinese and Thailand isolates, the structure and function of RGDV isolate, and viral transmission in South China, the RGDV segment S8, encoding the outer major capsid protein (Pns8) from five different locations in China, was sequenced and Pns8 of one isolate (XY) was analyzed in this study. To our knowledge, this is the first report on sequence analysis and genetic comparison of segment 8 from difference RGDV isolates, and the expression of the capsid protein.
HTML
-
Leaf samples of infected rice showing typical RGDV symptoms were collected from five Counties -XinYi (XY), GaoZhou (GZ), DeQin (DQ), BoLuo (BL), CongHua (CH) -in Guangdong Province, China. XY and GZ are neighbors, while the others are hundreds of kilometers away from each other. For control purposes, samples of uninfected plants were also collected. All samples were preserved at -80℃ until total RNA extraction.
Total RNAs of diseased or healthy leaves were extracted using a Total RNA Extraction (Plant Leaf) Kit (Watson, China), according to the manufacturer's instructions. Total RNAs were re-suspended in diethylpyrocarbonate (DEPC)-treated double distilled water and used as templates for RT-PCR.
The entire segment 8 (S8) of these five isolates were amplified respectively by RT-PCR using a one-step RT-PCR Kit (TaKaRa, China). The primers used were S8-P1 (5'-GGTATTTTTGTACCAACACG -3) and S8-P2 (5'-ATCATTTTTTGTGACCACAC-3') designed according to the S8 sequence of the Thailand isolate (TL) (GenBank Accession number: D13410). The detailed reaction conditions were as follows: reverse transcription at 50 for 30 min, pre ℃ denaturation at 94 fo ℃ r 5 min, followed by a standard PCR procedure with 30 cycles of denaturation at 94℃ for 1 min, annealing at 53 for 1 min, and extension ℃ at 72 for 2 min. A final extension was performed at ℃ 72 for 10 min. PCR products were analyzed on a ℃ 0.8% agarose gel.
-
Gel slices containing PCR products were excised from 0.8% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen, China). Purified PCR products were ligated into the pMD18-T Vector (TaKaRa, China) and recombined plasmids were transformed into E. coli competent cells. Cells carrying recombinant plasmids were selected on Luria–Bertani agar plates containing ampicillin, X-gal, and IPTG. Plasmid DNA was purified using the alkaline lysis protocol and sequenced using an ABI PRISM 3730 DNA analyzer (Applied Biosystems, USA). For each RGDV isolate, 3 or 4 independent clones originating from different RT-PCR reactions were selected.
-
Sequence data were compiled and analyzed using BioEdit (Isis Pharmaceuticals, Inc., Canada). The theoretical isoelectric point (pI) of the deduced peptide was predicted by the online tool "Compute pI/Mw Tool" at http://us.expasy.org/tools/pi_tool.html. Phylogenetic trees were constructed with MEGA (version 3.0) by using maximum parsimony analysis with 1000 bootstrap replicates. Sequence of Thailand isolate used for comparison and phylogenetic analysis in this study was obtained from the GenBank database (accession number: D13410).
-
RGDV segment 8 cDNA encoding a main capsid protein (Pns8) was PCR-amplified using a pair of primers 5'-GAATTCGATGTCGCGCCAAGCTTGG AT-3'and 5'-GTCGACTTAGTTTACTGTGTAATAC C-3' flanked by EcoRI and SalI restriction sites. The PCR product was subcloned into pET-28b (+) (Invitrogen, USA) at the EcoRI/SalI sites to yield an expression plasmid pET.CP. E. coli Rossetta (DE3)Ⅱ cells harboring pET.CP were incubated at 37 in a ℃ 100-mL flask containing 10 mL LB medium supplemented with 30 μg kanamycin/ml and 34 μg chloramphenicol/mL. When the OD600 reached 0.6, isopropylthio-α-D-galactopyranoside (IPTG) was added into the medium to a final concentration of 1 mmol/L, and E. coli Rossetta (DE3) Ⅱ cells were subsequently cultured at 42 to induce the protein expression. ℃ After 1, 3, 5, and 7 hours of culture respectively, the cells were harvested and total cell proteins were extracted and analyzed by SDS-PAGE.
Sample collection and preparation
Cloning and sequencing
Data Analysis
Prokaryotic expression of RGDV segment 8 cDNA (Pns8)
-
The genomic segment 8 of five isolates from China were amplified and sequenced successfully. The results showed that they were all 1578 bp in length and each contained one long open reading frame which extended for 1301 nt from 21 nt, and encoded the RGDV major outer capsid protein. The nucleotide sequence data reported in this paper were deposited in Genbank with accession numbers of AY999077 (BL), AY999078 (CH), AY999079 (DQ), AY999080 (GZ) and AY999081 (XY), respectively. The genomic segment 8 and the coding region of these five Chinese isolates shared 94.8% ~95.6% and 95.0%~96.0% nucleotide sequence identities with that of the Thailand isolate respectively, 97.3%~98.8% and 97.3%~99.1% within these five isolates respectively (Table 1). The variations of nucleotide sequence between Chinese isolates and the Thailand isolate were greater than that within the Chinese isolates. Variation seemed to be related to the geographic distance at the nucleotide level. But at the amino acid level, the situation changed. The deduced amino acid (Pns8) of GZ isolate was identical to that of the Thailand isolate, while the variability's of Pns8 within these five Chinese isolates ranged from 0.5% to 2.1% (Table 2). The residue variation position appeared to be random. It is of note that XY and GZ, the two neighbor isolates, still had a variation of 2 residues. The sequence divergence resulted in apparent changes in the theoretical isoelectric point (pI) of the deduced peptides. The pIs of the deduced peptides of both BL and DQ isolates were 7.62, while the others were 6.74. The effect of the changes remained unknown since no remarkable phenotype variability was observed within the five Chinese isolates.

Table 1. Comparison of the full-length and coding region sequences of genomic segment 8 of RGDV five Chinese isolates (BL, CH, DQ, GZ, XY) and Thailand isolate (TL)

Table 2. Divergence of the deduced amino acids of the outer main capsid protein gene of RGDV five Chinese isolates (BL, CH, DQ, GZ, XY) and Thailand isolate (TL)
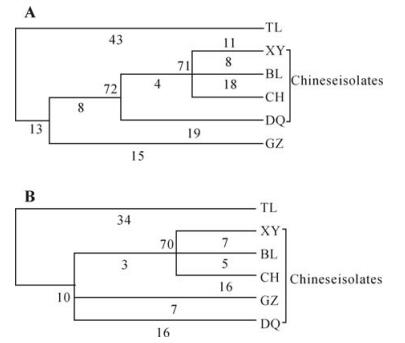
Two phylogenetic trees were built and shown in rectangular cladogram on the basis of the nucleotide sequences of the S8 and its coding region (Fig. 1). The two phylogenetic trees were similar and all the five Chinese isolates were grouped together, which indicated that they were more closely related to each other than to Thailand isolate. It is interesting that the two isolates, BL and CH, first reported in this paper, were clustered with XY isolate in one clade. This suggested that RGDV in BoLuo County and CongHua County might be newly introduced from Xinyi County, the original region, where the disease occurred first in China. The phylogenetic tree could not be constructed on the basis of amino acid sequences by maximum parsimony method because there were insufficient parsimony-informative sites.
-
Segment 8 cDNA encoding Pns8 of XY isolate was amplified and digested with EcoRI/SalI, ligated into linear vector pET-28b (+), and the recombinant pET.CP vector was obtained. Sequencing data showed that protein expression codons were completely correct. After E. coli Rossetta (DE3) Ⅱ cells containing pET.CP were grown at 37 ℃ and the OD600 reached 0.6, followed by further induction culture with IPTG for 3, 5, 7 hours at 42 respectively, the ℃ total cell fractions from growing 3-7 hours showed that the recombinant fusion protein was produced. This was further confirmed by SDS-PAGE analysis (Fig. 2). Molecular mass (51kDa) of the protein band was in good agreement with the size deduced from the whole amino acid sequence. On the contrary, no similar protein bands were detected in 3 control samples (Fig. 2). This preliminarily indicated that Pns8 was successfully expressed in E. coli Rossetta (DE3) Ⅱ.

Figure 2. Analysis of prokaryotic expression of recombinant RGDV pET.CP plasmid with SDS-PAGE. 1, E.coli. Rossetta(DE3) without induction; 2 Ⅱ, E.coli. Rossetta(DE3)Ⅱ induced with IPTG; 3, E.coli. Rossetta(DE3) containing Ⅱ pET.CP without induction; 4, Protein Marker; 5-8, E.coli. Rossetta (DE3) containing pET.CP was induced with IPTG Ⅱ for 1, 3, 5 and 7 hours, respectively.
Gene cloning and sequence analysis
Expression of Pns8 of RGDV XY isolate
-
RGDV has only been reported in China and in a few other Southeastern Asian Countries. Some previous reports have shown that there were some apparent differences in biological characteristics between the Chinese and Thailand isolates (2, 11), and revealed that there were different virus strains or strain differentiation in RGDV. Furthermore, RGDV was present in different regions in China, and could infect different rice cultivars. It is interesting therefore to determine whether these isolates are identical to each other. RGDV fragment S8, encoding the RGDV main outer coat protein, which might play some important roles in viral particle assembly, movement in host, transmission and the interaction between virus and host, was chosen for cloning and analyzing in this study.
Nucleotide sequence analysis showed that RGDV S8 shared 97.3%-98.8% identities within five Chinese isolates, and 94.8%-95.6% identities between the Chinese and Thailand isolates. These indicated that the genetic variation of RGDV S8 was more likely to be related to the geographic locations. However, the deduced amino acid sequences of the encoded protein (Pns8) showed a contradictory result. The Pns8 sequence of the GZ isolate from China was completely identical to that of the Thailand isolate, while the variability within five Chinese isolates ranged from 0.5% to 2.1%, and even a 2-residue change was found between two neighbor isolates (XY and GZ). This seemed to indicate that Pns8 amino acid sequence of RGDV was highly conserved, although there was higher variability over the entire nucleotide sequence of the S8 segment, which might be a consequence of adapting to the respective ecological environment in different geographical locations.
RGDV consists of 12 dsRNA segments. Only the nucleotide sequences of genome segments S2 (8), S3 (15), S5 (3), S8 (10), S9 (4), S10 (10), and S11 (9) from the Thailand isolate, and S9 from the Chinese XY isolate (1) have been reported. S8 sequence analysis indicated that RGDV S8 might not be sufficient to entirely determine the differences in biological characteristics between Chinese and Thailand isolates. Therefore, it is necessary to make an analysis and comparison of other RGDV segments between the isolates from two countries, which will be helpful to understand the relationship between different isolates of RGDV and the evolution of RGDV.
RGDV is restricted to plant phloem cells and has very low concentrations in rice plants, so it is difficult to get high purified yields from rice plants for preparation of antiserum (12). The protein expression in E. coli provides an alternative way to produce acid sequences between RGDV and RDV. enough protein in high quality for production of antiserum. Successful expression of the main outer capsid protein (Pns8) of RGDV in E. coli Rossetta (DE3) may provide a foundation for virus detection Ⅱ and analysis of virus functions.
Although different induction conditions were tested, Pns8 could not be expressed in all selected bacterial strains, such as E. coli BL21 (DE3), pLysS, BL21 (DE3) and JM109 (DE3). The protein could be detected in E. coli Rossetta (DE3)Ⅱ strain, though the expression amount was still low. This may be due to the existences of a number of rare codons (22.08%) of amino acids in Pns8 and several continuous and stable stem-loop structures in the translation initiation region of Pns8 mRNA (data not shown), which were not of benefit for the ribosome to contact and conjugate for enhancing translation start (5, 16). Suzuki et al. (14) propose that the E. coli expression system is not suitable for the protein expressions from the viruses of the family Reovirudae including RDV, WTV and RGDV. However, the main outer capsid protein from RDV was cloned into vector pTrcHisA and was efficiently expressed in E. coli (6). Lu et al. (7) cloned several segments of the second outer capsid protein gene of RDV into vector pTrcHisA, and found that the C-terminal segment could be expressed while the N-terminal segment could not. However, N-terminal segment could be successfully expressed if it was cloned into vector pET11d instead of pTrcHisA (7). These results indicated that the protein expression was significantly influenced by different expression strategies, selection of vectors and bacterial strains. Such situations will occur in the protein expression of RGDV, because there are high homologies of amino
Although Pns8 of RGDV in this study was expressed in the analysis of SDS-PAGE, it was not further confirmed with Western blot because of antiserum deficiency. Pns8 purification and preparation of the antiserum are currently under investigation.














 DownLoad:
DownLoad: