-
Simian virus 40 (SV40) is a member of the Polyomaviridae family which infects rhesus monkeys. In 1960,Sweet and Hilleman isolated SV40 from poliovirus vaccine produced in monkey kidney cells,and from rhesus monkey kidney cells (11). Soon afterwards,it was discovered that SV40 was able to induce tumors in newborn hamsters and transform many human cultured cell lines (3). Subsequently,SV40 was isolated from many human tumor tissues,including encephaloma,osteosarcoma and lymphoma (6, 9, 12). The SV40 genome can be sequenced by PCR from many human tumor tissues (5) ,suggesting that SV40 has pathogen representing an increasing risk to human health. Although it is uncertain whether SV40 directly induces these tumors,more and more data demonstrate that SV40 can transform cells by multiple cell signal transduction pathways via large T and small t proteins (1). To our knowledge,SV40 contamination of the polio vaccine during production and its close relationship with tumors has aroused scientific interest in conducting further studies.
The SV40 genome is a circular covalently closed dsDNA molecule with a length of about 5 200bp,consisting of a regulatory region,and early and late gene regions. The regulatory region promotes transcription and replication of the viral DNA. The early gene region encodes regulatory proteins for large T antigen (T-ag),small t antigen (t-ag) and early leader protein. The late gene region encodes structural proteins VP1,VP2,VP3 and agnoprotein,among which VP1 is the main structural protein,and VP2 and VP3 are the se-cond most important structural proteins. The agnoprotein is an SV40-specific protein in the Polyomaviri-dae family but its functions are not clear at the moment.
Despite the extreme conservation of the SV40 genome,recent studies indicate that heterogeneity exists in different isolates and is represented principally in the form of variations in the regulatory region and the C-terminus of the T antigen (4). In order to determine the epidemic SV40 genotype in Yunnan rhesus monkeys,data that is important for further studies,an SV40 strain was isolated from a captured Yunnan rhesus monkey. Its complete genome was sequenced and analyzed. Our findings demonstrate that this is a new virus isolate.
HTML
-
The virus was obtained from the isolated monkey kidney cell culture of a SV40 infected Yunnan rhesus monkey,and named as SV-IMB. The primes were synthesized and DNA was sequenced by Sangon Corporation. The Taq enzyme and clone vector PMD18-T were purchased from Takara Corporation.
-
The primers were designed based upon the complete sequence of the SV40 -776 strain.
-
The DNAs to be used as the template were abstracted via the conventional phenol-chloroform approach from cultures with obvious cellular patholo-gical changes. PCR amplification was performed on the virus genome by using the primers listed above subject to the follo-wing reaction program: 1cycle of denaturing at 95℃ for 1 min; 35 cycles of denaturing at 94℃ for 30s; annealing at 52℃ for 30s; 72℃ elongating at 72℃ for 45s; 1 elongating at 72℃ for 7min.The PCR products were cloned into a PMD-18 vector and transformed to E coli. DH5α after harvest and purification. Plasmids were abstracted from the screened clones,and the positive clones identified by PCR were sequenced by Sangon Corporation.
-
In order to analyze the homology and variations between SV-IMB strain and other 12 SV40 isolates,we compared their sequences by using the Clustal X 1.83 and Vector NTI 6.0 programs. The information on these isolates are presented in Table 2.The complete sequence of SV-IMB was submitted to the GenBank under accession number DQ660375.

Table 1. Primers for PCR Amplification

Table 2. History of SV40 isolates used in this study

Table 3. Nucleotide and amino acid homology comparison between SV-IMB and other SV40 isolates
1.1. Materials
1.2. Primers design
1.2. PCR amplification
1.3. Sequence analysis
-
The SV-IMB genome is composed of 5 246 nucleotides with 98.9% homology and 58 variable sites when compared to reference strain 776. The average homology was 98.1% as compared to 12 other SV40 isolates. In addition to the variations presented in the regulatory region and the C-terminus of T-Ag,amino acid variations were observed extending into the remaining conserved regions.
-
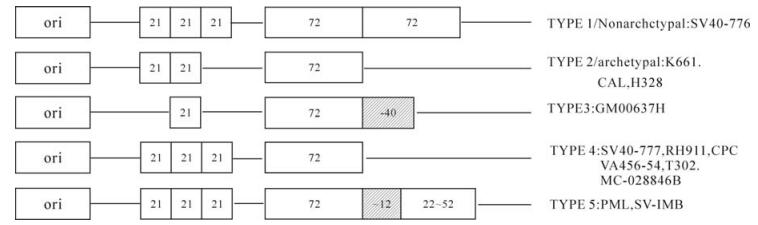
The regulatory region of the SV40-776 strain consists of a replication origin,three 21-bp repeat elements (G/C rich box) and two 72-bp enhancer elements and late promoter elements,termed non-archetypal type elements. Most of the isolates from naturally infected animals only consist of one 72-bp enhancer element,termed an archetypal element (7). Different isolates vary in the 21-bp repeat region and 72-bp enhancer region and can be categorized into 5 types (Fig. 1). The regulatory region of SV-IMB contains one 72-bp enhancer followed by another defective 53-bp enhancer,additionally,located in between these sites there is a 12-bp late promoter. This structure is similar in the PML isolate except that the defective enhancer only contains a 21-bp element-(CTAACTGACACAC) regardless of the different defective enhancer sequences. In addition,the same findings have been reported in many studies of the SV40 regulatory regions (2) ; however,the effect of the defective enhancers on virus replication is still unclear. Analysis of regulatory region sequences from the genomes of SV-IMB,GM00637H and PML/man etc.,revealed that replication of the late promoter is likely to stop during SV40 genome replication and that it would return back to a certain point in the enhancer region to initiate a new replication. In view of this,a stop at the 12-bp element in the late promoter region might be preferred,but there does not appear to be a definite location where new replication initiation occurs in an enhancer region.
-
It is noticed that nucleotide and amino acid homolo-gies were 98.9% when the SV-IMB isolate was compared to SV-776. Nucleotide variation led to the mutation of two amino acids,Y→F at amino acid 180 and P→S at amino acid 704 of the C-terminus. It is likely that such mutations lead to changes in the phosphorylation of the C-terminus of T antigen.
-
Although nucleotide variations in VP1 did not occur frequently,they led to the mutation of 4 amino acids which are characterized by an L→V at amino acid 73; G→V at amino acid 160; and YR→CS at amino acids 173 and 174. VP1 is the main capsid protein and is usually conserved. Only 9 of 366 amino acid sites vary among different SV40 isolates. As for SV-IMB,specific mutations occurred at 4 amino acids. Whether such mutations would lead to the variation of antigenic determinants needs to be explored further.
2.1. Complete genome sequence analysis
2.2. Sequence analysis in the regulatory region
2.3. Sequence analysis of large T antigen
2.4. Sequence analysis of VP1
-
A newly isolated SV-IMB from a Yunnan rhesus monkey is reported in this paper,and its complete genome was sequenced and analyzed. Our findings demonstrate that the genome of SV-IMB is extremely well conserved which is consistent with other SV40 isolates. Most variations are single nucleotide mutations except for the regulatory region.
In respect to sequencing findings,the regulatory region of SV-IMB is very well defined; it includes a complete 72-bp enhancer and a defective enhancer that is different from usual SV40 isolates. Xin Dang et al. reported the isolation of different SV40 variants from simian immunodeficiency virus-infected rhesus monkeys at different time points in different tissues. They performed sequence analysis on the regulatory regions of the screened clones. Their findings demonstrate that different types of regulatory regions might arise during in vivo replication of SV40 by an unknown mechanism. Continuous SV40 viral replica-tion is expected to result in a predominance of virus with TYPE 4 regulatory regions with three 21-bp repeats and one 72-bp enhancer,revealing the previous existence of such a regulatory region. TYPE 1 with 2 enhancers and TYPE 5 with one complete enhancer and one defective enhancer,are only rarely or not observed (2). Nevertheless,the type with only one enhancer is predominant in many diffe-rent SV40 isolates. As mentioned above,TYPE 4 with one enhancer predominates in the regulatory region,showing the highest possibility for survival,but such survival capabilities might be weakened when another enhancer is present. Indeed,the regulatory regions of SV40 isolates from naturally infected animals are most often TYPE 2 and TYPE 4 with only one enhancer due to selection pressure,while the regulatory regions of lab culture strains might exhibit types with no prior survival capabilities because there was not any selection pressure. Such interpretations seem to be reasonable; however,the regulatory region of the SV-IMB isolate from a naturally infected animal presented the latter characteristics. Whether this is due to the screening of a low numbers of clones or to more complicated reasons requires further investigation.
T-Ag has many functions,like mediating cell transformation,viral DNA replication,promotion and extension,regulating transcription of the virus genome,etc.,which are realized via binding to different DNA sequences and cytokines (10). It is reported that the C-terminus of the large T antigen has many phosphorylation sites and acetylation modification sites,and that the phosphorylation state of the C-terminus is closely associated with its ability to bind different DNAs. It is demonstrated that the DNA binding ability of T-Ag is enhanced,the level of viral DNA replication is increa-sed,and its transforming ability is lost when the Ser-677 and Ser-679 in the C-terminus are mutated to Ala that can not be phosphorylated (8). Assuming that the Ser-704 mutation of SV-IMB results in generating a new phosphorylation site,this mutation might influence viral replication and the cellular interactions of T-Ag.














 DownLoad:
DownLoad: