-
RNA silencing is a unique RNA-guided gene regulatory mechanism that operates in a wide range of eukaryotic organisms from plants to mammals. It compasses posttranscriptional gene silencing (PTGS) in plants as well as RNAi in animals. Common to both types of RNA silencing process is the production of ~21 nucleotide (nt) small RNA from structured or double strand RNA (dsRNA) by the endoribonuclease Dicer, an RNase enzyme. These siRNAs are incorporated into a multicomponent nuclease complex, the RNA-induced silencing complex (RISC) which can specially identify and degrade complementary target RNA in a homology-dependent manner, leading to posttranscriptional gene silencing (7). Recently, microRNA (miRNA), another type of small RNA which is derived from intrinsic hairpin RNA precursor, was also highlighted as an important type of RNAi. miRNAs often combine the target gene in the part of 3' untranslated region by in a complementary but inexact manner which attenuates the translational activity, and may also lead to degradation of target RNA (17).
RNAi is postulated to bean ancient immune mechanism used by cells to impede the actions of viruses, transgenes, and transposons. It plays an important role in defending invading pathogens while maintaining normal functions of cell development and apoptosis. Consistent with the notion that RNAi is a natural antiviral mechanism, miRNAs related to certain viruses in cells and siRNA derived from viruses in infection process were recently identified. Furthermore, some viruses were found to be able to encode proteins to suppress RNA silencing (9). Thus, viruses can antagonize the cell immune response at the gene level and enhance their ability to survive.
In this article, we designed short hairpin RNA (shRNA) targeted to the enhanced green fluorescence protein (EGFP) and luciferase genes expressed by vector in mammal cells and determined their abilities to down-regulate the target genes. Efficient screening systems of RNAi suppressor were established. Using this system, we further demonstrated the function of P19 of tomato bushy stunt virus to antagonist RNAi induced by shRNA in mammal cells.
HTML
-
Oligonucleotides were synthesized by Shanghai Bioasia Corporation. Sequences corresponding to the siRNA hairpin targets were as follows: shRNA-EGFP (5'-TCGAGGCTGACCCTGAAGTTCATCGAGTACTGGATGAACTTCAGGGTCAGCTTTTT-3')targeting the EGFP gene at sites 526nt-546nt, shRNA-Luc (5'-T CGAGAAGTGTTGTTCCATTCCATTTCAAGAGA ATGGAATGGAACAACACTTTTTTTTT-3')targeting the luciferase gene at sites 485nt-426nt, and the corresponding reverse sequences were also synthesized. After annealing, oligonucleotides were cloned into pTZU6+1 with SalⅠand XbaⅠrestriction sites. The constructs were identified by Sal Ⅰ digestion and further confirmed by DNA sequencing analysis.
-
pSG5mp19, which contains the DNA sequence of p19 and expresses the P19 protein in mammalian cells, was provided by Prof. Charle (Institut de Biologie Moleculaire des Plantes, France). To detect the protein of P19 expediently, we constructed a plasmid expressing the P19 fused with a his tag. We obtained the DNA sequence from pSG5mp19 by PCR with the primers 5'-AGTCTCGAGACCATGGAACGAGCTA T-3' and 5'-GACGGATCCCTCGCTTTCTTTTTCGA-3'. The sequence was then inserted into the pcDAN-3.1-myc-his (-) between restriction sites Xho Ⅰ and BamH I. After transfecting the plasmids into HepG2 cells, the mRNA expression of P19 and P19-his were confirmed by RT-PCR. Human glyceraldehydes-3-phosphate dehydrogenase (hGAPDH) was detected at the same time as a positive control. Primers for hGAP-DH were 5'-GGCTCTCCAGAACATCAT-3 and 5'-CA CCTGGTGCTCAGTGTA-3'. The protein of P19-his was also confirmed by the immunofluorescent method. The rabbit antibody pointing to the his tag was purchased from santan cruz. The second antibody labeled with FITC was from Beijing Zhongshan Corporation.
-
HepG2 cells were cultured at 37℃, 5% CO2 in medium 1640 supplemented with 10% FBS. pEGFP-N1 (promega) was transfected into cells with lipofectamine (invitrogen) according to the manufacturer's instructions. 24hr after transfection, G418 (500 μg/ml) was added into the culture medium, and the green fluorescence of cells were observed by fluorescence microscopy every day. Cells producing strong green fluorescence were harvested and individually seeded into 96 well plates, guaranteeing only one cell in each well. Cells producing strong green fluores-cence were amplified. To confirm the integration of the GFP gene into the cell genome, the genomic DNA of HepG2 cells which expressing GFP was extracted by the phenol-chloroform method and then used as the template for PCR to detect GFP. The primers used in this test were as follows: forward primer (5'-GATGG TACCCTATGGTGAGCAAGGGC-3'), reverse primer (5'-GACAGTACTGCTTGTACAGCTCGTCCA-3'). The genome DNA of hepG2 was used as a control.
-
To study the effect of P19 on the GFP RNAi system, 4 groups of various plasmids were transfected by lipofectamine into the cells of HepG2-GFP as follows: 1) pSG5mp19+pshRNA-GFP; 2) pcDNAp19-his+psh-RNA-GFP; 3) pcDNA3.1-myc-his(-) +pshRNA-GFP; 4) pcDNA3.1-myc-his(-)+pshRNA-Luc. To analyze the influence of P19 on luciferase RNAi system, 4 groups of different plasmids were transfected into cells as follows: 1) pSG5mp19+pshRNA-Luc; 2) pcDNAp19-his+pshRNA-Luc; 3) pcDNA3.1-mychis(-) +pshRNA-LUC; 4) pcDNA3.1-myc-his(-)+pshRNA-GFP. In each group, pGL3 and pRL TK were also transfected. The former was used as reporting gene and the latter was used to normalize the transfection efficiency.
-
The green fluorescence of different groups was observed by fluorescence microscopy every day posttransfection. At 72hr post-transfection, proteins of cell lysis were separated by SDS-PAGE and transferred by electroblotting onto a polyvinyllidene difluoride membrane. A rabbit monoclonal antibody directed against eGFP (BD) was used and identified by a second HRP-conjugated antibody (Beijing Zhongshan) through enhanced chemiluminescence (Amersham). Signals were detected with genesnap and quantified with the genetool software. At the same time, actin protein was detected as a control in the same manner with a goat antibody directing actin and a second antibody directing goat IgG (both from Beijing Zhongshan).
-
Total RNA was extracted from cultured cells posttransfection with the RNAeasy kit (Qiagen) and then the RNA was digested with DNaseⅠto exclude DNA contamination. To quantify the RNA from EGFP, the hGAPDH was amplified at the same time as a control. Primers for EGFP in the tests were as follows: forward (5'-GCAGCACGACTTCTTCAA -3'), reverse (5'-GT CCATG CCGAGAGTGAT-3'). The PCR products were analyzed by gel electrophoresis and the band was quantified with the genetools software.
-
48h after transfection, cells were lysed by 1× luciferase passive lysis buffer (Promega) and centrifuged at 12 000g for 15 sec, the liquid was used to detect luciferase activity by a multi-function enzyme analysizer (Gene corp). The relative activity of firefly luciferase was counted by normalizing to renal luciferase. The reporter values represented averages±1 SD from at least three independent transfections.
pshRNA constructions
Plasmid expressing P19 fused with his tag
Cell line stably expressing GFP
plasmids transfection
Western blotting
Semi-quantative RT-PCR
Luciferase assay
-
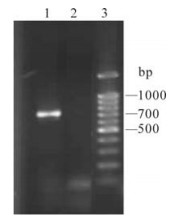
To construct a RNA interference model in mammal cells, we established the cell line stably expressing GFP (the cell line was named HepG2-GFP) by G418 selection culture. We obtained a cell line with strong green fluorescence observed by fluorescence microscopy after one-month culture. The insertion of EGFP into genome DNA was then confirmed by PCR. We detected the fragment of EGFP with the genome DNA as the template (Fig. 1. lane 1) whereas the same fragment did not appear in the control test (Fig. 1).
-
The tomato bushy stunt virus is a type of plant virus. To confirm its expression in mammal cells, we detected at the mRNA and protein levels by different methods. The mRNA of P19 were detected in cells of HepG2-GFP and HepG2 transfected with the Psg5mp19 or pcDNAp19-his respectively by RT-PCR. We furthermore detected the P19-his protein by an antibody targeting the his tag using the immunofluenscent method and P19-his proteins were observed to be primarily located in the plasma.
-
We next designed the siRNA targeting EGFP to suppress its expression. Here, a vector expressing strategy and a vector with RNAⅢ promoter (U6) was chosen. The vector pTZU6+1 can drive the transcription of short hairpin RNA precisely, which would be transferred into a functional type of siRNA in mammal cells by the Dicer. When the HepG2-GFP was transfected with the plasmid pshRNA-GFP, the fluorescence was reduced significantly compared with the control group, which was transfected with the plasmid pshRNA-Luc (Fig. 2.A). This was consistent with the results from western blotting (Fig. 2.B). Analysis with the Genetools software indicated the amount of GFP decreased by 70%. To determine the influence on mRNA levels, we further detected mRNA by a semi-quantative RT-PCR test and found that pshRNA-GFP lead a decrease in mRNA level of EGFP (Fig. 2.C) by 78%. Therefore it could be concluded that the shRNA down-regulated the expression of EGFP and the down-regulation was a consequence of the degradation of mRNA. On the basis of the successful RNAi system described above, we studied the ability of P19 to suppress the RNAi effect in mammalian cells. When P19 and shRNAGFP were co-expressed in HepG2.GFP, we observed the phenol menon that the fluorescence recovered to a significant intensity compared with those cells without P19. The effect on efficiency on GFP expression was further evaluated by western blot for protein levels and by RT-PCR for mRNA levels. In these tests, P19 recovered the GFP protein expression as well as mRNA expression (Fig. 3) increasing the expression of GFP by 80% and mRNA levels by 70% compared to the controls.
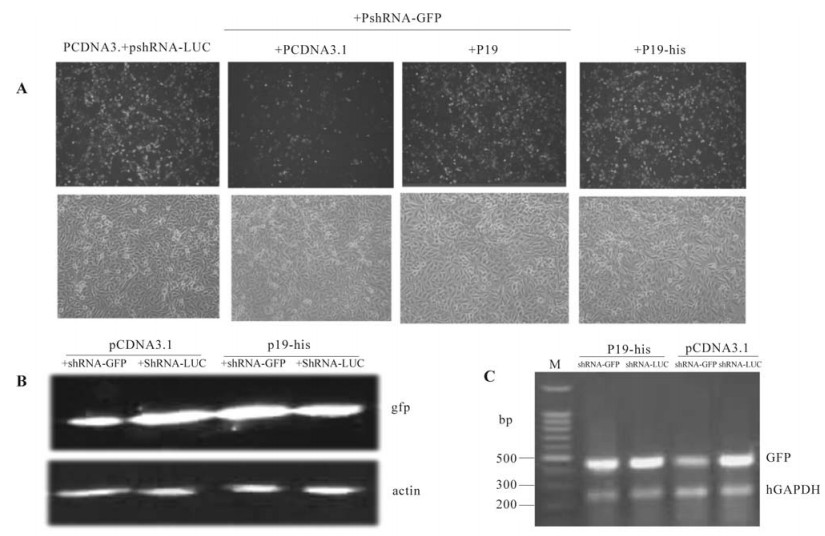
Figure 2. P19 counteract the effect of shRNA on GFP expression. A: fluorescence observed by fluorescent microscope. ShRNA-GFP down-regulate fluorescence intensity whereas the P19 recovered the fluorescence in hepG2.GFP. B: The GFP levels were observed decreased by the shRNA-GFP and increased when P19 was introduced. C: mRNA was found decreased when shRNA existed whereas P19 recovered the mRNA level of GFP.
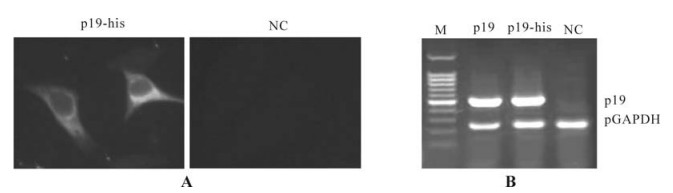
Figure 3. P19 expressed in mammal cells detected in the level of protein and mRNA. A: P19-his was observed by immunnofluorescent method in hepG2 cells transfected with pCDNAp19-his. Cells transfected with pCDNA3.1-myc-his was used as negative control (NC). P19-his was observed located in the plasma mostly. B: mRNA of P19 was also detected in the cells transfected with pSG5mp19 or pCDNAp19-his, respectively. mRNA extracted from Cells transfected with pCDNA3.1-myc-his was used as negative control. A fragment of hGAPDH gene was amplified in all three samples while fragment of P19 was only amplified from cells transfected with pSG5mp19 and pCDNAp19-his.
-
To further understand the shRNA interference efficiency, we designed siRNA to target firefly luciferase. When the cells were transfected with pshRNA-Luc and the reporter vector, pGL3 as control (Promega), which expresses firefly luciferase under the control of SV40 promoter, the luciferase activity reduced by 70% (Fig. 4) compared with the control group. It showed that the shRNA-Luc designed could down-regulate the expression of luciferase gene efficiently. When it was studied by the luciferase RNAi system, P19 was also found to be able to recover the luciferase activity significantly. Compared with the control group, when P19 was introduced into the cells, the relative luciferase activity increased to about 80%. The his tag did not impair the function of P19. Results from the luciferase RNAi system coincided with those from the GFP RNAi system.
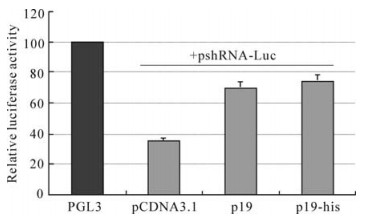
Figure 4. P19 rescued the luciferase expression in the RNAi system. The relative luciferase activity was counted by firefly luciferase activity devided by rena luciferase activity and the value of control group was standardized to 100. when shRNA-Luc was introduced, the firefly luciferase decreased significantly. When P19 was added into the RNAi system, luciferase activity recovered to a high level.
The cell line of HepG2-GFP expressing GFP stably
P19 and P19-his expressed in mammal cells
P19 recovered the expression of GFP down-regulated by shRNA
P19 rescued the expression of luciferase in RNAi system
-
RNAi is an ancient immune surveillance mechanism on gene level. It was shown to act as an efficient antiviral system in plant and insect cells and might also played an antiviral role in mammal cells (2, 11). To counteract the antiviral effect of RNAi and enhance their existing ability, many plant and insect viruses express different RNAi suppressor proteins (14). These proteins always play important roles in the virus infection process and are important pathogens (12). The first identified RNAi suppressor, HC-pro of tobacco etch potyvirus (TEV), was found when researchers studied the coinfection phenomenon in plants (8). Later, some other RNAi suppressors encoded by plant viruses were discovered and the mechanism of RNAi inhibition became better understood (15). Furthermore, several animal viruses such as flock house virus, influenza virus and reovirus were also found to encode proteins having the same effect as an RNAi suppressor (19, 10). Recently, HIV-1 and PFV-1 [primate foamy virus type 1, a retrovirus similar to HIV] were found to be able to produce such RNAi suppressors too (1, 3). Interestingly, HIV-1 can produce a siRNA in the infected cells to downregulate its Env expression while a cellular miRNA was verified to target the sequence of PFV-1 and could restrict the accumulation of PFV-1. These reports indicated that RNAi mechanism may also play an important role in vertebrate cells and the RNAi suppressor exists as an counteraction strategy to this antiviral mechanism.
Similar to the phenomenon of RNAi, RNAi suppressor was firstly studied in the field of plant research. Nowadays, we have known that RNAi suppressors could take effect at different steps in the RNAi pathway (14). HC-pro, δ3 factor of reovirus, and NS1 of the influenza virus countact RNAi by binding long dsRNA and reduce production of siRNA. Tat of HIV-1 can also limit the production of siRNA by influencing the activities of Dicer. Some suppressors such as P19 can bind the siRNAs and prohibit them into RISC (4). Other suppressors may also act at various steps. For example, some of them may influence the activities of members of RISC, and some of them may limit the transduction of systematic silencing signals in cells (18). In summary, although much of the mechanism of RNAi suppressing is still to be studied, we can concluded that different suppressors may have different interference methods and show different abilities to inhibit RNA silence.
To date, miRNA was regarded as a kind of siRNA and considered part of the RNAi process by some researchers although single strain miRNA has some differences to short double strain RNA. miRNAs were also produced by Dicer and incorporated into RISC at last. miRNA and siRNA crossed partly in their pathway at least. If a protein could influence RNAi, it may also influence miRNAs. Since miRNAs have very important roles in keeping normal development and normal biological activity of cells, cells would be influenced when miRNA levels were changed. Patrice D et al studied the influence on miRNAs of several suppressors and discovered that most of them showed an obvious effect and could produce abnormalities (13). We could also predict the pathogenesis of the suppressor of animal viruses by influencing miRNA function. For example, a persistant production of the SRS in chronic virus infection may help to produce tumor and other chronic diseases.
Tomato bushy stunt virus is a 4.7k nt plus RNA virus which infect agriculture plants and herbs. The P19 is essential for the viral pathogenity since it can enhance the ability of the virus to survive in infected plants by counteracting the RNAi system. It has been established that the RNAi mechanism is the most important defence strategy in plants. Daniel et al firstly reported that P19 could inhibit the RNAi effect in plants and pointed out it could combine the siRNAs and prohibit it incorporate into RISC. Recently, Ye et al elaborated the crystal structure of P19 and explained the physiochemical basis of its ability to combine siRNAs (5). In this report, RNAi systems targeting GFP and luciferase gene were constructed and used to identify the RNAi suppressor characteristics of P19. The results showed that P19 could also suppress RNAi effects induced by short hairpin RNAs in mammal cells as well as suppressing RNAi induced by synthetic siRNA or long dsRNA in plants. Our research confirmed that the suppressing ability of P19 was non-sequence specific for it suppressed both the RNAi targeting GFP and the RNAi targeting luciferase. Furthermore, our study showed that the RNAi system in mammal cells induced by vector derived shRNA is suitable for screening RNAi suppressors, and could be a more efficient approach compared with the methods such as transgenic plant models or virus infection models.
Scientists attached much importance to therapy exploitation of RNAi in viral infection diseases and cancer diseases when synthetic siRNAs were found to be able to down-regulate the homologous gene expression by activating the RNAi mechanism in mammal cells (6). Today, much improvement has been achieved in this field (16). But the discovery of RNAi suppressors encoded by viruses will bring some new questions to the application of siRNA drugs. Benasser pointed out that HIV could resist the persistent vector-derived shRNAs since its Tas was a RNAi suppressor (1). It may also be the case that some mammal viruses such as HCV, HBV and SARS-coV may also encode such RNAi suppressors and these factors could influence the therapy strategy of RNAi. There are still many unknown factors to be studied in this field. The knowledge of RNAi suppressor will not only enrich our understanding of RNAi phenomenon and interaction between virus and host but also help the exploitation of RNAi as a therapy strategy.














 DownLoad:
DownLoad: