HTML
-
Noroviruses remain an important cause of acute non-bacterial gastroenteritis in all age groups worldwide, and are mainly associated with viral foodborne outbreaks (Hall et al., 2013). The viruses cause an estimated 200, 000 deaths and 1, 000, 000 hospitalizations in children under five years of age in developing countries annually (Patel et al., 2008). Noroviruses belong to the family Caliciviridae, and are non-enveloped, positive sense, single-stranded RNA viruses (Green, 2013). Frequent change of the viral genome contributes to the high ge-netic diversity of norovirus strains (Bull et al., 2007; Bok et al., 2009). Noroviruses are classified into seven genogroups (GI-GVII), of which GI and GII are major causes of acute gastroenteritis in humans (Vinje, 2015). The viruses are highly infectious and transmitted through the fecal-oral route by person-to-person contact and consumption of contaminated food or water (Mathijs et al., 2012).
Since an appropriate tissue culture system for human noroviruses is lacking, molecular techniques including RT-PCR, RT-nested PCR, and real-time RT-PCR have been employed to detect noroviruses in clinical and environmental samples (Lees et al, 2010; Pang and Lee, 2015; Kittigul et al., 2016). Among those techniques, real-time RT-PCR has been widely used to detect and quantify norovirus genomes since this assay has a high sensitivity and specificity, provides fast results, and decreases the risk of carry-over contamination (Baert et al., 2007; Neesanant et al., 2013; Yan et al., 2013; Fuentes et al., 2014; Farkas et al., 2015). Nevertheless, the sensitivity and specificity of real-time RT-PCR methods vary due to the highly diversified norovirus genomes and real-time RT-PCR protocols which utilize different primers, probes, reagents, and conditions (Kageyama et al., 2003; Mattison et al., 2011; Schultz et al., 2011). Various commercial quantitative real-time RT-PCR assays are available in the market (Butot et al., 2010; Dunbar et al., 2014; Hyun et al., 2014), although commercial real-time RT-PCR assays revealed a lower detection rate of noroviruses and could not detect a majority of norovirus GI strains as compared with the European international assay (Butot et al., 2010). The present study aimed to evaluate the performance of three real-time RT-PCR assays based on the TaqMan method (a commercial real-time RT-PCR and two in-house real-time RT-PCR assays) for the detection and quantification of noroviruses GI and GII.
-
Noroviruses GI and GII DNA positive controls (1 × 107 DNA copies/mL) supplied in the Norovirus Real Time RT-PCR kit (Shanghai ZJ Bio-Tech, Shanghai, China) according to the European Authorized Representative Obelis S.A.(Brussels, Belgium) were used to determine the sensitivity of three real-time RT-PCR assays for detection of noroviruses GI and GII. Sixty one archived fecal samples (18 GI and 43 GII) which were positive for noroviruses by RT-nested PCR (Kittigul et al., 2010) were used to evaluate the real-time RT-PCR assays. Group A rotavirus DNA positive control, hepatitis A virus DNA positive control, and poliovirus positive control were used for testing specificity of the assays. A total of 40 archived fecal samples, which were positive for each 10 rotavirus, norovirus GI and norovirus GII, and 10 rotavirus/ norovirus-negative fecal samples, were also employed to test the specificity amongst norovirus genogroups.
-
Viral RNA was extracted from 140 μL of fecal sample diluted 1:10 in 0.05 mol/L phosphate-buffered saline using the QIAamp® viral RNA extraction kit (QIAGEN Gmbh, Hilden, Germany), following the manufacturer's instruction. RNA was tested directly for norovirus by real-time RT-PCR or stored at -80 ℃ until use.
-
The three real-time RT-PCR assays were carried out using different RT-PCR reagents and thermocycling conditions. All assays were performed in separate tubes for norovirus GI and norovirus GII. A total of 5 µL of RNA was added to 15 µL of real-time RT-PCR master mix and amplified in a LightCycler 2.0 instrument (Roche Diagnostics, Mannheim, Germany).
Assay A was a commercial Norovirus Real Time RT-PCR kit (Shanghai ZJ Bio-Tech, Shanghai, China) containing specific primers, TaqMan probes labelled with 6-carboxyfluorescein (FAM), norovirus GI or GII Super Mix and RT-PCR Enzyme Mix for the simultaneous detection of noroviruses GI and GII. The cycling conditions consisted of reverse transcription at 45 ℃ for 10 min; initial denaturation at 95 ℃ for 15 min; then 45 cycles of denaturation at 95 ℃ for 5 s and annealing/extension at 60 ℃ for 30 s.
Assay B used the LightCycler RNA Master HybProbe reagent (Roche Diagnostics, GmbH Mannheim, Germany) containing specific primers and TaqMan probes targeted to the highly conserved ORF1-ORF2 junction to amplify noroviruses, namely primers COG1F, COG1R and probes RING1a-TP, RING1b-TP for norovirus GI and primers COG2F, COG2R and probe RING2-TP for norovirus GII (Kageyama et al., 2003). All TaqMan probes were labelled with FAM at 5′ end and 6-carboxy-tetramethylrhodamine (TAMRA) at 3′ end of the oligonucleotides. The master mix was composed of 1 × LightCycler® RNA Master Hybprobe containing Tth DNA polymerase, reaction buffer, and dNTP mix (with dUTP instead of dTTP), 3.25 mmol/L Mn (OAc)2, 0.4 µmol/L of each primer for norovirus GI or GII, 0.2 µmol/L of each probe for norovirus GI or GII and PCR grade water. The thermocycling profiles included reverse transcription at 58 ℃ for 30 min; initial denaturation at 95 ℃ for 4 min; then 45 cycles of denaturation at 95 ℃ for 15 s and annealing/extension at 56 ℃ for 1 min.
Assay C employed the RealTime ready RNA Virus Master reagent (Roche Diagnostics, GmbH Mannheim, Germany) with the same specific primers and TaqMan probes as used in assay B. The master mix consisted of 1× Reaction buffer containing RT-PCR Reaction buffer, dNTP mix (with dUTP instead of dTTP), and MgCl2, 0.4 µmol/L of each primer for norovirus GI or GII, 0.2 µmol/L of each probe for norovirus GI or GII, 1× Enzyme Blend (Transcriptor RT, and Taq DNA polymerase), and PCR grade water. The cycling conditions were reverse transcription at 50 ℃ for 10 min; initial denaturation at 95 ℃ for 4 min; then 45 cycles of denaturation at 95 ℃ for 15 s, annealing at 56 ℃ for 30 s, and extension at 72 ℃ for 15 s.
-
For sensitivity determination of assays A-C, the reactions were performed with 3-4 repetitions on different days using 10-fold serial dilutions of norovirus GI or GII DNA positive controls at starting concentrations of 1 × 107 DNA copies/mL or 2.5 × 104 DNA copies/reaction, respectively. The positive results were determined as the quantification cycle (Cq) values of ≤ 38. The sensitivity of each assay was the lowest concentration of norovirus GI or GII giving a positive result by real-time RT-PCR.
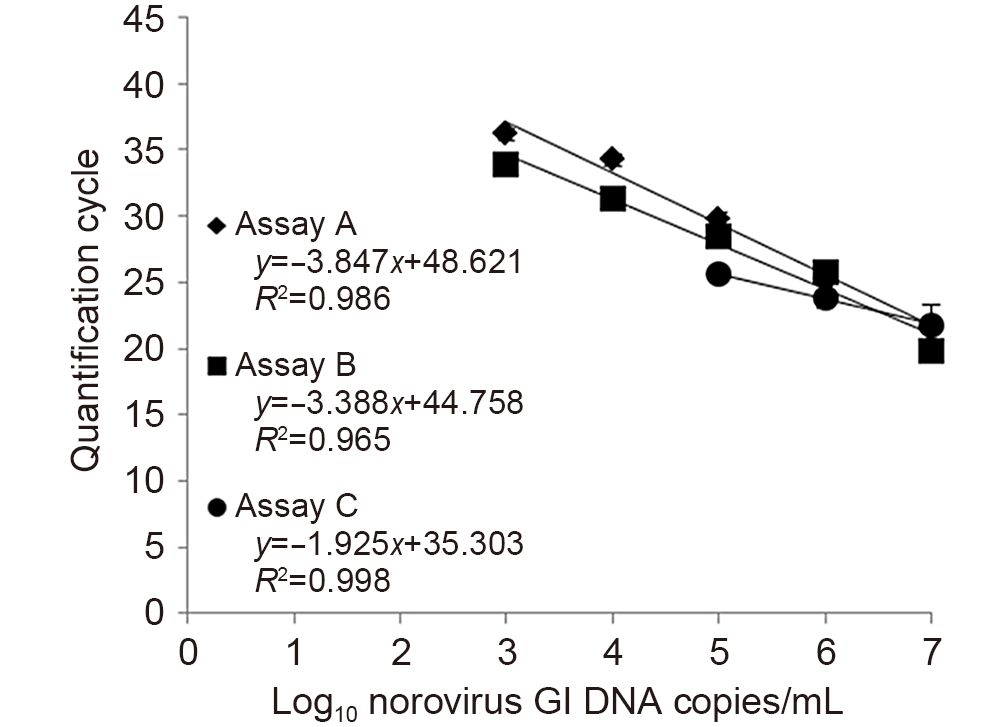
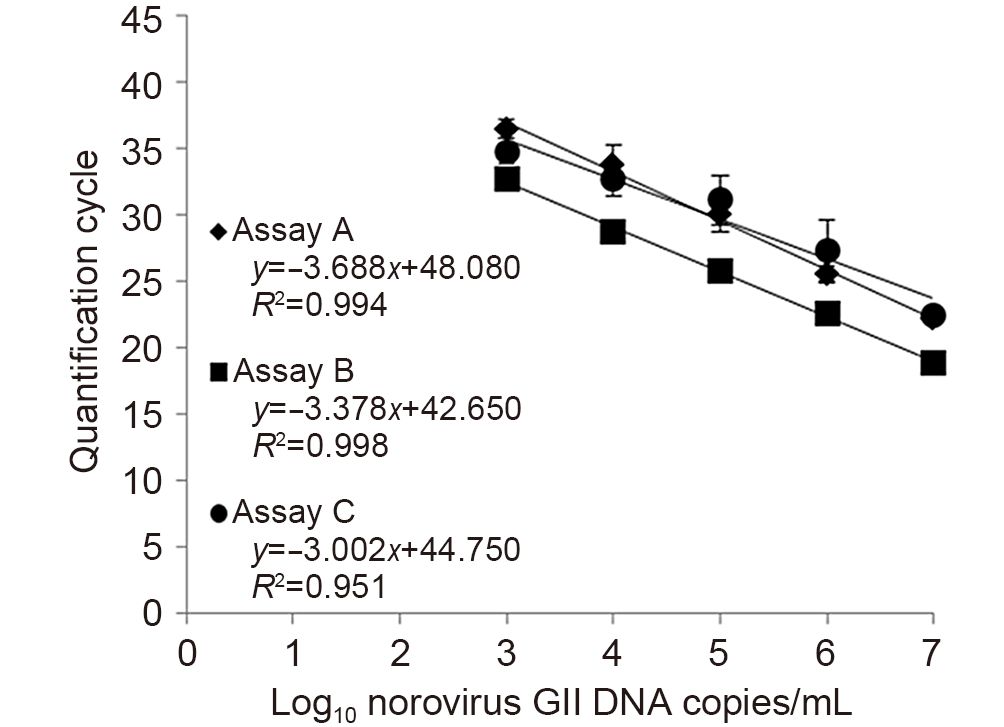
To evaluate the amplification efficiency of the real-time RT-PCR assays, standard curves of noroviruses GI and GII DNA copy numbers (1 × 107, 1 × 106, 1 × 105, 1 × 104 and 1 × 103 DNA copies/mL) versus Cq values were generated. The mean Cq values in repeated experiments were plotted to determine the slope and regression coefficient (R2) values. Subsequently, the amplification efficiency was calculated using the following equation: efficiency (E)= 10-1/slope-1(Bustin et al., 2009).
DNA positive controls of rotavirus and hepatitis A virus, and RNA extracted from poliovirus positive control, rotavirus-positive and rotavirus/ norovirus-negative fecal samples were tested using specific primers, probes, and thermocycling conditions for noroviruses GI and GII to determine the specificity of the three real-time RT-PCR assays. Additionally, norovirus GI-and GII-positive samples were tested to determine the specificity amongst norovirus genogroups.
-
To evaluate the performance of the three real-time RT-PCR assays, RNA was extracted from norovirus GI-and GII-positive fecal samples and analyzed using assays A-C. The detection rates, Cq values, and DNA copy numbers of norovirus genomes obtained from the three different real-time RT-PCR assays were compared.
-
For qualitative determination of the detection of noro-viruses in fecal samples, Cochran Q's test was used to test significant differences amongst the three real-time RT-PCR assays. For quantitative determination, the Cq values and the DNA copy numbers of noroviruses obtained from the real-time RT-PCR assays were compared using the Friedman test. A P-value of < 0.05 was considered to be statistically significant. Statistical anal-ysis was performed using the Statistical Package for the Social Science (SPSS) version 18.
Norovirus positive controls and fecal samples
Viral RNA extraction
Real-time RT-PCR assays
Analytical sensitivity, amplification efficiency, and specificity
Comparison of three real-time RT-PCR assays for norovirus detection
Statistical analysis
-
The analytical sensitivities of assay A (Norovirus Real Time RT-PCR kit), assay B (LightCycler RNA Master Hybprobe), and assay C (RealTime ready RNA Virus Master) were determined using 10-fold serial dilutions of noroviruses GI and GII DNA positive controls. For norovirus GI, the lowest DNA copy number that could be detected by assays A and B was 1 × 103 DNA copies/mL (2.5 DNA copies/reaction), whereas assay C was only able to detect a higher copy number of 1 × 105 DNA copies/mL (2.5 × 102 DNA copies/reaction), as shown in Table 1. For norovirus GII, the sensitivities of the three assays were equal (1 × 103 DNA copies/mL) but assay B gave a lower Cq value of 31.89 ± 1.08(mean ± SD)(Table 2).
Norovirus GI, DNA copies/mL Assay Aa Assay Ba Assay Ca Mean Cq ± SDb Mean Cq ± SDb Mean Cq ± SDb 1 × 106 25.26 ± 0.23 25.72 ± 0.36 23.85 ± 0.86 1 × 105 29.58 ± 0.54 28.75 ± 0.16 25.63 ± 0.01 1 × 104 34.20 ± 0.39 31.77 ± 0.36 – 1 × 103 36.11 ± 0.48 35.06 ± 0.74 ND 1 × 102 – – ND Notes: –, negative; ND, not done. aAssay A: Norovirus Real Time RT-PCR kit; Assay B: LightCycler RNA Master HybProbe; Assay C: RealTime ready RNA Virus Master; b3–4 repeat experiments. Table 1. The analytical sensitivities of three real-time RT-PCR assays for the detection of norovirus GI
Norovirus GII, DNA copies/mL Assay Aa Assay Ba Assay Ca Mean Cq ± SDb Mean Cq ± SDb Mean Cq ± SDb 1 × 105 30.22 ± 0.93 27.09 ± 1.50 31.15 ± 1.89 1 × 104 34.24 ± 1.26 29.08 ± 0.78 32.99 ± 1.11 1 × 103 36.13 ± 0.87 31.89 ± 1.08 34.64 ± 0.89 1 × 102 – – – Notes: –, negative. aAssay A: Norovirus Real Time RT-PCR kit; Assay B: LightCycler RNA Master HybProbe; Assay C: RealTime ready RNA Virus Master; b3–4 repeat experiments. Table 2. The analytical sensitivities of three real-time RT-PCR assays for the detection of norovirus GII
-
To evaluate the amplification efficiency of the real-time RT-PCR assays, the mean Cq values of each assay were plotted against log of DNA copy numbers of the noro-virus positive controls. For norovirus GI, assays A and B generated a 5-log range of linearity (1 × 103-1 × 107 DNA copies/mL) while assay C only generated a 3-log range of linearity (1 × 105-1 × 107 DNA copies/mL), as shown in Figure 1. The amplification efficiency of assay B was 97.31% with a slope of-3.388 and an R2 of 0.965 demonstrating a higher efficiency than the other two assays. Assays A and C gave efficiencies of 81.95%(slope =-3.847, R2 = 0.986) and 230.74%(slope =-1.925, R2 = 0.998), respectively. For norovirus GII, all assays generated a 5-log range of linearity (Figure 2). The calculated efficiency of assay B was 97.71% with a slope of-3.378 and an R2 of 0.998 exhibiting higher efficiency than that of assays A and C at 86.70%(slope =-3.688, R2 = 0.994) and 115.33%(slope =-3.002, R2 = 0.951), respectively.
-
The specificities of the three real-time RT-PCR assays for detection of noroviruses GI and GII were assessed using known positive controls and fecal samples of other enteric viruses. All real-time RT-PCR assays gave nega-tive results for the detection of noroviruses GI and GII in the positive controls of rotavirus (1 × 106 DNA copies/mL), hepatitis A virus (1 × 107 DNA copies/mL), and po-liovirus (7.14 × 102 50% tissue culture infective dose [TCID50]/mL), rotavirus-positive fecal samples (6.69 × 106-1.45 × 109 DNA copies/mL), and rotavirus/ norovirus-negative fecal samples. Those assays also showed negative results for the detection of norovirus GII in norovirus GI-positive fecal samples (1.03-1.83 × 104 DNA copies/mL) and for the detection of norovirus GI in norovirus GII-positive fecal samples (1.79 × 103-2.11 × 109 DNA copies/mL). No cross-reaction with any of the other enteric viruses examined was observed.
-
The three real-time RT-PCR assays were compared for the detection of noroviruses GI and GII in fecal samples. Of 61 samples, 51(83.6%) showed corresponding results (positive or negative by all assays), while 10(16.4%) showed discrepant results. The agreement rate between assays A and B (56/61, 91.8%) was higher than between assays B and C (55/61, 90.2%) and assays A and C (52/61; 85.3%).
Of 18 norovirus GI-positive fecal samples, 5(27.8%) were detected by assay A at a higher frequency than assays B (16.7%) and C (5.6%). However, there was not a statistically significant difference. The DNA copy numbers of norovirus GI determined by all three assays showed the same range of 104-106 DNA copies/mL (Table 3). Of 43 norovirus GII-positive fecal samples, 39(90.7%) were detected by assay B at a higher frequency than assays A (88.4%) and C (81.4%). This difference was not statistically significant. Obviously, the mean Cq value of assay B was lower than assays A and C with statistical significance (P-value, 0.000). The mean DNA copy numbers of norovirus GII determined by all three assays were not different, and showed the same range of 102-109 DNA copies/mL (Table 4).
Assaya No. of positive samples/total (%) Quantification cycle (Cq) Noroviurs GI, DNA copies/mL Mean Range Mean Range A 5/18 (27.8) 29.58 22.17–31.88 2.00 × 106 1.03 × 104–9.33 × 106 B 3/18 (16.7) 28.44 26.92–29.70 4.08 × 105 1.93 × 104–1.17 × 106 C 1/18 (5.6) 21.55 21.55 1.85 × 106 1.85 × 106 P-value 0.050b –c –c Notes: a: Assay A: Norovirus Real Time RT-PCR kit; Assay B: LightCycler RNA Master HybProbe method; Assay C: RealTime ready RNA Virus Master method; b: Cochran's Q test; c: Statistical analysis was not determined because Cq and DNA copies/mL of Method C were obtained from one norovirus GI-positive sample (i.e. there is no mean Cq or DNA copies/mL value). Table 3. The detection and quantification of norovirus GI in fecal samples using three real-time RT-PCR assays
Assaya No. of positive samples/total (%) Quantification cycle (Cq) Noroviurs GII, DNA copies/mL Mean Range Mean Range A 38/43 (88.4) 28.38 13.04–35.36 7.27 × 107 4.56 × 102–2.11 × 109 B 39/43 (90.7) 25.79 15.67–30.20 4.83× 107 7.29 × 102–1.24 × 109 C 35/43 (81.4) 28.06 13.77–34.25 1.21× 108 2.20 × 102–3.47 × 109 P-value 0.368b 0.000c 0.846c Notes: a: Assay A: Norovirus Real Time RT-PCR kit; Assay B: LightCycler RNA Master HybProbe method; Assay C: RealTime ready RNA Virus Master method; b: Cochran's Q test; c: Friedman test. Table 4. The detection and quantification of norovirus GII in fecal samples using three real-time RT-PCR assays
Amongst 61 archived norovirus-positive fecal samples, 34 were identified previously for norovirus genotype (Kittigul et al., 2010). The three real-time RT-PCR assays could detect a variety of noroviruses including GI.2, GII.2, GII.3, GII.4(2006a and 2006b variants), GII.6, GII.12, GII.17, and GII.21. Assay A was able to detect GI.6, whereas the other two assays could not. Overall, assay A could detect norovirus genotypes with a frequency equal to assay B (85.3%) and higher than assay C (76.5%), as shown in Table 5.
Norovirus genotype No. of samples Assay Aa Assay Ba Assay Ca GI.2 1 1 1 1 GI.6 2 2 – – GII.2 2 1 1 1 GII.3 3 2 2 2 GII.4 2006a 2 2 2 2 GII.4 2006b 13 11 12 10 GII.6 2 2 2 2 GII.12 1 1 1 1 GII.17 1 1 1 1 GII.21 7 6 7 6 Total (%) 34 29 (85.3) 29 (85.3) 26 (76.5) Notes: a: Assay A: Norovirus Real Time RT-PCR kit; Assay B: LightCycler RNA Master HybProbe; Assay C: RealTime ready RNA Virus Master. Table 5. Norovirus genotypes detected by three real-time RT-PCR assays
The analytical sensitivities of real-time RT-PCR assays
The efficiencies of real-time RT-PCR assays
The specificities of the real-time RT-PCR assays
Comparison of three real-time RT-PCR assays for detection of noroviruses in fecal samples
-
Noroviruses have been recognized as an important cause of acute gastroenteritis outbreaks in developed and developing countries (Patel, 2008). Because human noro-viruses are not cultivable, real-time RT-PCR has become the method of choice for laboratory diagnosis and identification of noroviruses in clinical and environ-mental samples (Stals et al., 2013; Pang and Lee, 2015). Variations in the sensitivity and specificity of real-time RT-PCR procedures have been reported with differences in primers, probes, reagents, and conditions (Kageyama et al., 2003; Mattison et al., 2011; Schultz et al., 2011). The available commercial quantitative real-time RT-PCR kits and real-time RT-PCR reagents are still expensive in developing countries. The analytical sensitivity of assays A and B for the detection of norovirus GI DNA positive control was 2.5 DNA copies/reaction and higher than that of assay C (250 DNA copies/reaction). However, the three real-time RT-PCR assays gave an equal sensi-tivity of 2.5 DNA copies/reaction for the detection of the norovirus GII DNA positive control. These results are comparable with previous studies that have determined the sensitivity of real-time RT-PCR at 10-50 genome copies/reaction for norovirus GI and 1-300 genome copies/reaction for norovirus GII (Kageyama et al., 2003; Mattison et al., 2011; Schultz et al., 2011). Assay B showed a higher efficiency (97%) for the real-time RT-PCR detection of both norovirus genogroups than the other two assays. The amplification efficiency should be in the range of 90%-110% and the low reaction efficiency ( < 90%) found in assay A may be a consequence of contamination with Taq inhibitors, non-optimal annealing temperature, old or inactive Taq polymerase, poorly designed primers, or amplicons with secondary structures. The high reaction efficiency ( > 110%) seen in assay C may be a consequence of the formation of primer-dimers or non-specific amplicons (Taylor et al., 2010). Assay B utilizes Tth DNA polymerase which is different from assay C which uses Transcriptor reverse transcriptase and Taq DNA polymerase. Tth DNA polymerase possesses both reverse transcriptase and polymerase activity and is at least 100-fold more efficient than Taq DNA polymerase in RT-PCR (Myers and gelfand, 1991). Additionally, this enzyme is more resistant to PCR inhibitors, and viral nucleic acids can be amplified in a single step by RT-PCR (Poddar et al., 1998). The variability of sensitivity and efficiency of real-time RT-PCR assays can be influenced by primers/probes, real-time RT-PCR reagents and thermocycling conditions (Espy et al., 2006).
The current study used norovirus GI and GII DNA positive controls for assessment of real-time RT-PCR sensitivity and efficiency, and generation of standard curves for quantification of noroviruses in clinical samples. To compare the performance of the three real-time RT-PCR assays, RNA transcript positive controls would be more appropriate because the differences in reverse-transcription steps could affect the sensitivity and efficiency of the assays. We also performed the experiments using RNA transcripts determined by assay B and the lower sensitivities of norovirus GI (106 RNA copies/mL) and norovirus GII (104 RNA copies/mL) were obtained (data not shown). It is recommended the use of either double-stranded DNA or the actual viral genome of hepatitis A virus (HAV) for the construction of the standard curve. HAV RNA gave the similar slope of the standard curve but the intercept value of RNA was significantly higher (Costafreda et al., 2006). DNA positive controls as well as RNA transcripts were utilized for the deter-mination of the sensitivity of real-time RT-PCR for nor-oviruses (Kageyama et al., 2003; Mattison et al., 2011; Schultz et al., 2011). More intensive studies need to be done to address this concern.
The three real-time RT-PCR assays showed no cross-reactivity with other enteric viruses and amongst noro-virus genogroups demonstrating high specificity of the primers and probes used in this study. This finding is consistent with a previous study in which cross-reaction between norovirus GI and norovirus GII was not observed (Kageyama et al., 2003). Nevertheless, the evaluation of norovirus real-time RT-PCR procedures from participating Canadian laboratories exhibited cross-reaction of norovirus GII with rotavirus and sapovirus in sensitive detection (Mattison et al., 2011).
The performance evaluation of the three real-time RT-PCR assays could be seen clearly on the use of clinical samples. For the testing of norovirus-positive fecal sam-ples, assay A was in agreement with assay B in terms of corresponding positive or negative results. Assay A detected norovirus GI at the highest frequency. The primers/probes for norovirus GI published by Kageyama et al.(2003) and used in assays B and C were shown to be less sensitive for the detection of norovirus GI in clinical and environmental samples (Loisy et al., 2005; Van Stelten et al., 2011). The redesigning the primers/probes specific for norovirus GI and optimizing for real-time RT-PCR could improve the sensitivity and amplification efficiency of the assay (Van Stelten et al., 2011). However, the limitation of this study is the small numbers of norovirus GI-positive fecal samples as determined by real-time RT-PCR. The complexity of fecal material might affect the PCR results, therefore, the detection of norovirus GI in fecal samples needs to be further studied. For norovirus GII, assay B provided the highest detection rate. Although the frequency of norovirus GII-posi-tive fecal samples determined by assay B is not much different from assay A, the lowest mean Cq value of assay B demonstrates the highest efficiency of DNA amplification indicating the highest performance of the assay B to amplify norovirus GII in clinical samples.
The three real-time RT-PCR assays could detect a variety of known norovirus genotypes including GI.2, GII.2, GII.3, GII.4 2006a variant, GII.4 2006b variant, GII.6, GII.12, GII.17, and GII.21. All but one genotype (GII.21) detected by the real-time RT-PCR assays correspond to the findings of a previous study (Butot et al., 2010). The norovirus GII.4 2006b variant caused global outbreaks during 2006-2007(Siebenga et al, 2009). GII.17 emerged and caused outbreaks of gastroenteritis in China (Lu et al., 2016), whereas GII.21 was prevalent in sporadic cases with acute gastroenteritis in Thailand (Kittigul et al., 2010). Regarding the cost of the real-time RT-PCR, assay A is approximately 18 USD/test, assay B 15 USD/test, and assay C 12 USD/test. With respect to sensitivity, specificity, and efficiency of the real-time RT-PCR assays, assay B is the in-house method suggesting to be employed for the detection and quantification of noroviruses GI and GII due to the good performance characteristics of the test and the reasonable cost. Rapid detection and quantification of noroviruses will be useful for gastroenteritis outbreak investigations, disease surveillance, and health risk analysis of norovirus infection.
-
This work was supported by research grant from the Thailand Research Fund (TRF) through the Royal Golden Jubilee Ph.D. program (Grant No. PHD/0085/2554), and the Thai Government Budget through Mahidol Uni-versity, fiscal year 2015-2017. The authors thank The Language Center, Faculty of Graduate Studies, Mahidol University for editorial assistance.
-
All the authors declare that they have no competing interests. The study was approved by the Ethical Review Committee for Human Research, Faculty of Public Health, Mahidol University. The protocol complied with a "Research with Exemption" category.
-
KDR performed the experiments and analyzed the data. PCP and PPD participated in the experiments. LRK designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.














 DownLoad:
DownLoad: