-
Dear Editor,
The genus Orbivirus, within the family Reoviridae, includes 22 virus species (Kinget al., 2011). They are distributed globally, but are particularly prevalent in Europe, Asia, and Africa. In addition, they can be transmitted by ticks or other hematophagous insect vectors, including Culicoides, mosquitoes, and sandflies (Belaganahalliet al., 2015).
Orbiviruses contain double-stranded RNA as their genetic material, which includes 10 segments (S1–S10) of various lengths. Owing to this segmented structure, genetic reassortment occurs frequently among orbiviruses. Reassortment occurs when different segmented viruses of the same vector type infect a single host cell, and ge-nomic segments of the "parental" viruses are exchanged and repackaged during progeny formation (Simon-loriere and Holmes, 2011). Reassortment can result in novel viral genotypes and subsequent phenotypes, which can alter the mechanism of immune escape, the potential range of hosts or vectors, and the mechanism of virulence or pathogenicity (Mcdonald and Patton, 2011). In general, reassortment represents a critical mechanism in the evolu-tion of segmented viruses. Accordingly, to understand viral diversity, it is important to identify and characterize reassortant taxa.
To identify novel reassortant orbiviruses, specimens were collected during the active mosquito season (from June to September) in Xishuangbanna, Yunnan Province in southwestern China in 2007. A sample was virus-positive if it had a cytopathic effect in three successive cell passages (Zuoet al., 2014). Supernatants containing the virus were identified by reverse transcription-polymerase chain reaction (RT-PCR) with genus-or species-specific primers designed for potential mosquito-borne viruses (Leiet al., 2014). Virus-containing samples that could not be identified by RT-PCR were identified by high-throughput sequencing (HTS) (Patel and Jain, 2012). Complete nucleotide sequences for segments 1–10 (Seg1–10) of a virus, referred to as Banna orbivirus (BAOV), were determined. The sequences have been submitted to GenBank, with sequential accession numbers KX455487–KX455496. The full-length sequence genome map and the protein sequence of the 10 BAOV segments are shown in Figure 1A.
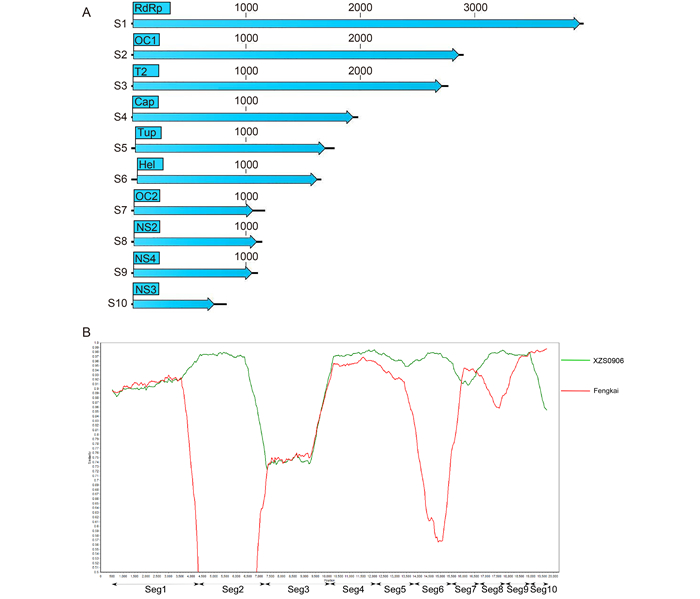
Figure 1. (A) The full-length sequence genome map and protein sequence of 10 BAOV segments. (B) Similarity among BAOV, Fengkai orbivirus, and XZ0906. The red line represents TIBOV (XZ0906); the green line represents Fengkai orbivirus; the horizontal axis indicates the degree of sequence similarity; the vertical axis indicates the position of genomic S1–10 sequences.
Recently, several novel orbiviruses have been isolated. Among the most common orbiviruses, Tibet orbivirus (TIBOV) (XZ0906) was first isolated from a pool of Anopheles maculatus mosquitoes in Tibet, China in 2009. TIBOV (YN12246) was isolated from Culicoides spp. in Yunnan, China, and Fengkai orbivirus, which also belongs to TIBOV, was isolated from a pool of Culex fatigan mosquitoes in Guangdong Province, China in 2008. A comparison of gene and protein sequences of BAOV segments to those of Fengkai orbivirus and XZ0906 is summarized in Table 1. The complete ge-nomes of BAOV, Fengkai orbivirus, and XZ0906 were 19270, 19349, and 19235 bp, respectively, and had similar GC contents–42.4%, 42.7%, and 42.5%, respectively. Each genomic segment encodes a single major open reading frame. S1–10 encode seven structural proteins: RNA-dependent RNA polymerase (RdRp), outer capsid protein one (OC1), inner sub-core shell 'T2' protein, Cap, Tup, Hel, and OC2, and three non-structural proteins (NS), NS2, NS4, and NS3 (Mosset al., 1992).
Virus/segment Segment length (bp) Protein encoded Predicted protein length (aa) ORFs (bp, including stop codon) 5′ NCRs 3′ NCRs %GC Content Accession No. BAOV Seg1 3950 RdRp 1326 12-3926 11 23 41.1 KX455487 Seg2 2901 OC1 966 13-2865 12 35 39.8 KX455488 Seg3 2769 T2 916 11-2716 10 52 43.8 KX455489 Seg4 1978 Cap 654 9-1940 8 37 42.9 KX455490 Seg5 1775 TuP 546 83-1696 82 78 44.3 KX455491 Seg6 1656 Hel 505 47-1626 46 29 45.2 KX455492 Seg7 1165 OC2 355 18-1067 17 97 41.9 KX455493 Seg8 1142 NS2 365 21-1100 20 31 40.9 KX455494 Seg9 1102 NS4 352 15-1057 14 44 42.4 KX455495 Seg10 832 NS3 241 13-726 12 105 44.9 KX455496 Total 19270 42.4 KX455497 Fengkai Seg1 3950 RdRp 1326 12-3926 11 23 40.5 NC_027803.1 Seg2 2999 OC1 941 14-2791 13 207 40.0 NC_027804.1 Seg3 2761 T2 914 18-2717 17 43 44.0 NC_027805.1 Seg4 1979 Cap 654 9-1940 8 38 43.0 NC_027806.1 Seg5 1775 TuP 563 32-1696 31 78 43.1 NC_027807.1 Seg6 1636 Hel 535 27-1607 26 28 42.0 NC_027811.1 Seg7 1165 OC2 355 18-1067 17 97 45.0 NC_027812.1 Seg8 1151 NS2 368 21-1109 20 41 42.0 NC_027813.1 Seg9 1100 NS4 352 15-1055 14 44 44.2 NC_027814.1 Seg10 833 NS3 238 22-726 21 106 41.5 NC_027815.1 Total 19349 42.7 XZ0906 Seg1 3950 RdRp 1326 12-3926 11 23 40.8 KF746187.1 Seg2 2888 OC1 962 14-2854 13 33 40.7 KF746188.1 Seg3 2769 T2 914 18-2717 17 51 44.2 KF746189.1 Seg4 1978 CaP 654 9-1940 8 37 42.8 KF746190.1 Seg5 1775 TuP 563 32-1696 31 78 43.4 KF746191.1 Seg6 1636 Hel 535 27-1607 26 28 42.1 KF746192.1 Seg7 1165 OC2 355 18-1067 17 97 45.3 KF746193.1 Seg8 1142 NS2 365 21-1100 20 41 43.6 KF746194.1 Seg9 1100 NS4 352 15-1055 14 44 44.6 KF746195.1 Seg10 832 NS3 238 22-726 21 105 41.3 KF746196.1 Total 19235 42.5 Table 1. Characteristics of dsRNA genome segments and proteins of BAOV, Fengkai orbivirus, and XZ0906
To determine the phylogenetic relationships among BAOV and other viruses in the genus Orbivirus, multiple sequence alignments were generated using Mega v6.06 with the complete sequences of BAOV and 15 other orbiviruses (Tamuraet al., 2013) (Supplementary Table S1). Phylogenetic trees based on all 10 viral genome segments were constructed from the nucleotide sequence alignments by using the maximum likelihood method with the GTR+G+I model. The robustness of the tree was established by a bootstrap analysis with 1, 000 replicates. To determine similarity, Simplot 3.5 (http://www.med.jhu.edu/deptmed/sray/download/; provided by S. Ray) was used to generate similarity plots based on the Cluster alignment (Mosset al., 1992). To better understand the genetic relationships among BAOV and other orbiviruses, 10 phylogenetic trees were constructed based on S1–10 (Supplementary Figure 1A–1J). BAOV, Fengkai orbivirus, and XZ0906 were grouped into a single cluster, which was distinct from other orbiviruses. S3 from BAOV had a higher amino acid sequence homology with S3 from YN12246 (94%) than with S3 from Fengkai orbivirus (80%) or S3 from XZ0906 (80%). However, sequences for only two segments (S1 and S3) from YN12246 are available in the GenBank database. The remaining BAOV segments had high identities to those of Fengkai orbivirus and XZ0906. S2 and S6 from BAOV exhibited high sequence similarities with S2 and S6 from Fengkai orbivirus. S10 from BAOV exhibited high sequence similarity with S10 from XZ0906 (Supplementary Figure 1A–Supplementary Figure S1). The degrees of nucleotide and amino acid sequence homology between the 10 segments in BAOV and those of other orbiviruses were also examined using BLASTn and BLASTp, respectively (Supplementary Table S2). S2 and S6 from BAOV had higher nucleotide and amino acid sequence homology with S2 and S6 from Fengkai orbivirus (97% and 98%) than with S2 and S6 from Tibet orbivirus (70% and 71%). S10 from BAOV had higher nucleotide and amino acid sequence homology with S10 from XZ0906 (99%) than with S10 from Fengkai orbivirus (85%). Simplot and Bootscan analyses revealed the similarity among BAOV, Fengkai orbivirus, and XZ0906 genomic sequences in each region (Figure 1B). These results are consistent with those of the BLASTn analysis, as shown in Supplementary Table S2. S2 and S6 from BAOV were more similar to S2 and S6 from Fengkai orbivirus than to S2 and S6 from XZ0906. S10 from BAOV was more similar to S10 from XZ0906 than to S10 from Fengkai orbivirus.
Reassortment is a common event in the evolution of viruses. In this study, we report a novel mosquito-borne reassortant Orbivirus species, which we named Banna orbivirus (BAOV). The virus was isolated from a Culex tritaeniorhynchus pool collected in Xishuangbanna, Yunnan Province, China in 2007. Whole genome and phylogenetic analyses revealed that BAOV is a reassortant of several Tibet orbivirus strains. It has a double-stranded RNA genome of 19, 270 bp, containing ten segments (S1–S10) of various lengths. The 10 segments of BAOV had high nucleotide and amino acid sequence similarity with segments of either Fengkai orbivirus or Tibet orbivirus (TIBOV, XZ0906 strain). Phylogenetic analyses indicated that the novel BAOV is a reassortant derived from XZ0906 and Fengkai orbiviruses, and is a Tibet orbivirus. This is the first report of a novel genomic reassortment of Tibet orbivirus isolated in China; these findings contribute to our understanding of the diversity of orbiviruses.
HTML
-
This research was supported by a grant from the China Mega-Project on Infectious Disease Prevention (grant numbers 2013ZX10004-605, 2013ZX10004-607, 2013ZX10004-217, and 2011ZX10004-001), the National Hi-Tech Research and Development (863) Program of China (grant numbers 2014AA020108, 2012AA022-003), and the National Natural Science Foundation of China (grant numbers 81273138, 81572045). The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
Supplementary figure/tables are available on the websites of Virolo-gica Sinica: www.virosin.org;link.springer.com/journal/12250.
-
Species Abbreviation Strain/serotype Collection Region year Segment accession No. African horse sickness virus AHSV HS29-62/serotype 1 1968 South Africa KP009770.1-KP009779.1 Blue tongue virus BTV SZ97-1/serotype 1 1993 Southern India JN848759-JN848768 Epizootic hemorrhagic disease virus EHDV New Jersey/serotype1 1994 USA KU173875.1-KU173883.1 Equine encephalosis virus EEV HS103-06 2008 Israel AB811630.1-AB811639.1 Eubenangee virus EUBV AUS1963/01 2012 United Kingdom JQ070376.1-JQ070385.1 Great Island virus GIV CanAr-42 2010 United Kingdom ADM88592-ADM88601 Kemerovo virus KEMV EgAn 1169-61 2012 Russia KC288130.1-KC288139.1 Tribec virus TRBV TRBV 2010 Germany HQ266581.1-HQ266590.1 Palyam virus PALV Chuzan 2015 China KT887180.1-KT887189.1 Peruvian horse sickness virus PHSV PHSV 1997 Peru NC_007748.1-NC_007757.1 St Croix River virus SCRV SCRV 1999 France AAG34363-AAG34372 Umatilla virus UMAV USA1969/01 2010 United Kingdom AEE98368-AEE98377 Yunnan orbivirus YUOV YUOV 2004 France NC_007656.1-NC_007665.1 Tibet orbivirus TIBOV Fengkai 2008 China KR822286.1-KR822295.1 XZ0906 2009 China KF746187.1-KF746196.1 Table S1. Summary of all virus strains used in this study
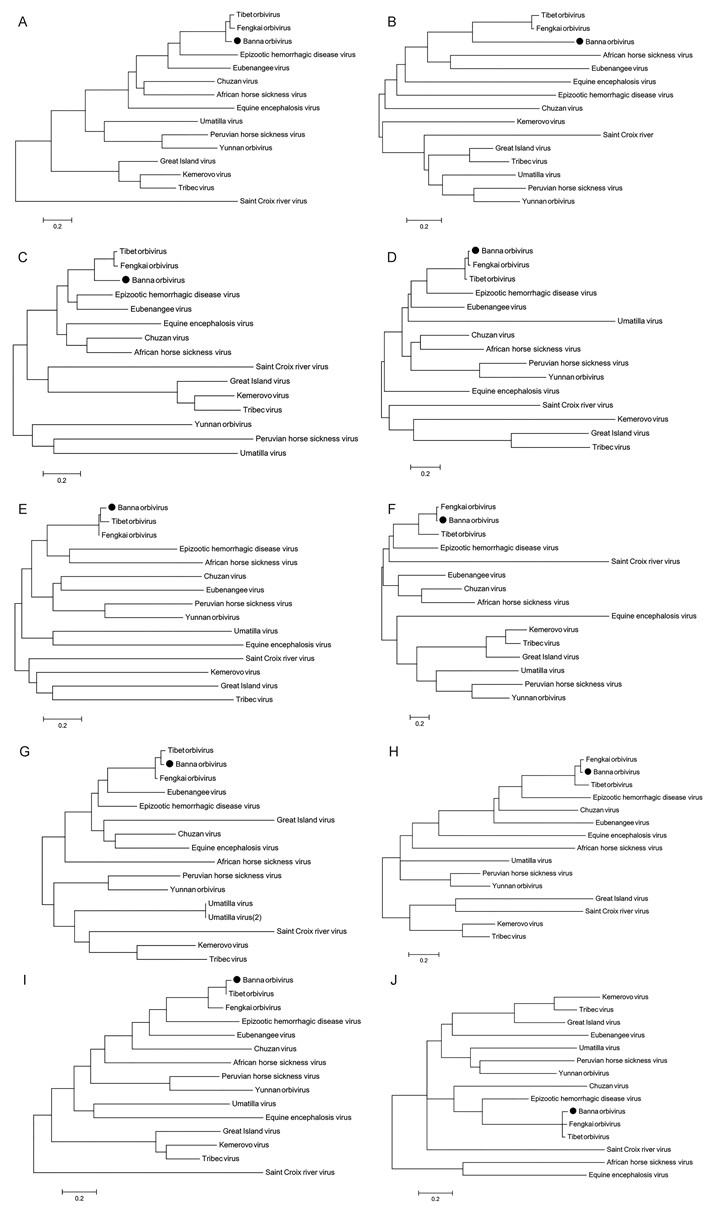
Figure S1. Phylogenetic analyses of nucleotide sequences of the 10 genomic segments (S1–S10) of BAOV and other known orbiviruses. The accession numbers of the other strains are shown in Table S1
Region TIBOV (XZ0906) Fengkai orbivirus nt aa nt aa Cov (%) Iden (%) Cov (%) Iden (%) Cov (%) Iden (%) Cov (%) Iden (%) Seg1 100 92 100 99 100 91 100 99 Seg2 12 70 99 42 95 97 97 98 Seg3 100 80 99 96 99 80 99 96 Seg4 100 96 100 99 100 98 100 99 Seg5 100 96 100 99 99 97 100 99 Seg6 96 71 98 85 96 98 98 100 Seg7 100 95 100 99 100 93 100 99 Seg8 100 89 100 97 100 97 100 98 Seg9 100 97 100 92 100 97 100 92 Seg10 100 99 98 99 100 85 98 94 Table S2. The organization and the BLAST results for different regions of BAOV genome
















 DownLoad:
DownLoad: