-
Dear Editor,
A common reason for drug failure during long-term treatment of chronic hepatitis B with nucleot(s)ide analogues (NUCs) is the emergence of drug resistance (Das et al., 2001). Most primary NUCs-resistant mutations identified in clinical samples have been limited to a minority of amino acids (usually less than three kinds) that replace the wild type amino acid at a single site (Kirishima et al., 2002). However, each site should, theoretically, have the same opportunity to be mutated to any of the 19 amino acids other than that of the wild type during the DNA synthesis catalyzed by the error-prone viral polymerase (Bartholomeusz et al., 2004). Therefore, precisely how these undiscovered possible mutations influence the viral phenotype is an interesting research question. To address this issue, Baldick et al. constructed a comprehensive panel of clones containing every possible amino acid at the three primary amino acid positions related to entecavir resistance (rtT184, rtS202, or rtM250) in the lamivudine (LAM)-resistant hepatitis B virus (HBV) background (rtM204V + rtL180M) to determine the phenotypic impact of these substitutions (Baldick et al., 2008). Although extensive mutagenesis analysis is very useful, this kind of assay is usually difficult to conduct. The construction of mutated plasmids would be laborious if there are a large amount of mutants need to be tested. In addition, the transfection step can be a source of variation, and the plasmid DNA transfected into cells, if not completely eliminated during the purification step, will interfere with the detection of HBV DNA replicative intermediates. Herein, we report an alternative method for conducting an extensive mutagenesis phenotype assay of HBV polymerase. This method is based on our previously developed polymerase trans-complement strategy (Luo et al., 2016). Using the well-known LAM-resistant mutation rtM204 site as a target, we have demonstrated the feasibility of this method.
The alternative strategy to perform an extensive mutagenesis phenotype assay that combines randomized mutagenesis and HBV polymerase trans-complementation is outlined in Figure 1A. The reverse primer Rpol204 (Supplementary Table 1), which has two randomized nucleotides (NNT) at the second and third sites of the rt204 codon (ATG), was used to amplify a fragment together with a forward primer targeting the backbone of the plasmid pLentipol-D. The fragment was inserted at the corresponding region on pLentipol-D by fragment-substitution reaction (FSR) (Hu et al., 2012), generating plasmids carrying NN mutations at the rt204 codon. Plasmids carrying different mutations were pooled and used to package the lentiviruses. After the transduction of the lentiviruses carrying the blasticidin-resistant gene into the cell line 293HBV-pol-, monoclonal cells were selected with antibiotics. The genomic DNA samples from these clones were amplified with a pair of primers (the forward primer anneals to a region upstream of rt204M, and the reverse primer anneals to a sequence on the lentiviral vector) and sequenced to identify the mutations of rt204 in each clone. Finally, expanded monoclonal cells were used to test the replication capability of the HBV DNA and the LAM susceptibility by real-time PCR.
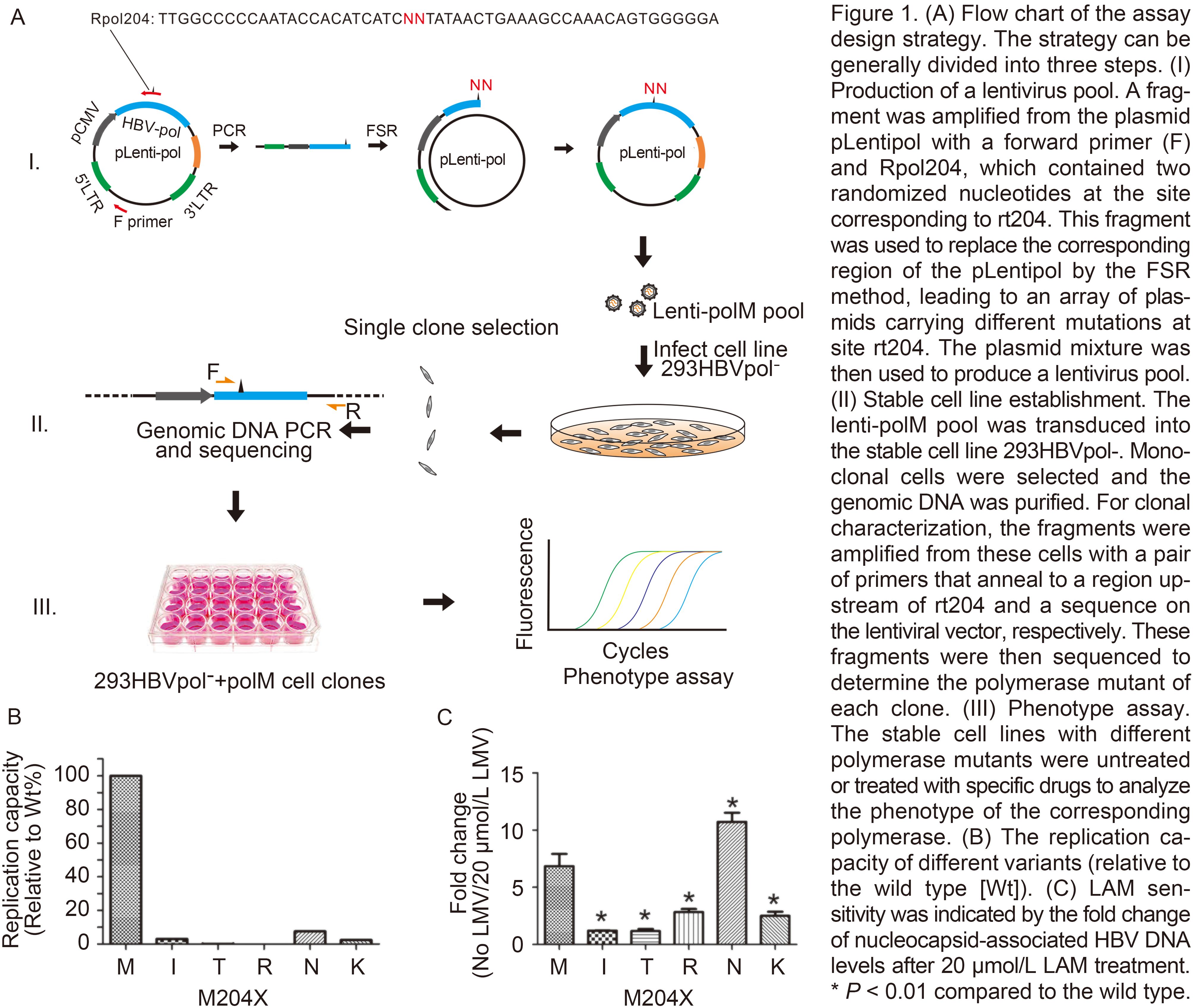
Figure 1. (A) Flow chart of the assay design strategy. The strategy can be generally divided into three steps. (I) Production of a lentivirus pool. A fragment was amplified from the plasmid pLentipol with a forward primer (F) and Rpol204, which contained two randomized nucleotides at the site corresponding to rt204. This fragment was used to replace the corresponding region of the pLentipol by the FSR method, leading to an array of plasmids carrying different mutations at site rt204. The plasmid mixture was then used to produce a lentivirus pool. (II) Stable cell line establishment. The lenti-polM pool was transduced into the stable cell line 293HBVpol-. Monoclonal cells were selected and the genomic DNA was purified For clonal characterization, the fragments were amplified from these cells with a pair of primers that anneal to a region upstream of rt204 and a sequence on the lentiviral vector, respectively. These fragments were then sequenced to determine the polymerase mutant of each clone. (III) Phenotype assay. The stable cell lines with different polymerase mutants were untreated or treated with specific dmgs to analyze the phenotype of the corresponding polymerase. (B) The replication capacity of different variants (relative to the wild type [Wt]). (C) LAM sensitivity was indicated by the fold change of nucleocapsid-associated HBV DNA levels after 20 pmol/L LAM treatment. * P < 0.01 compared to the wild type.
Nucleotides (ANN) Amino acid Number of clones AUG* M204 WT 3 AUA* M204I 4 AUU 1 AUC 2 ACA M204T 0 ACU 2 ACC* 7 AAA M204K 5 AAG* 2 AAU* M204N 3 AAC 4 AGA M204R 3 AGG* 1 AGC M204S 0 AGU 0 37 Note: The genomic DNAs were purified from 40 clones, 37 of which were found to have the integrated polymerase gene. The number of different mutants is listed. * Cell lines with these mutants were subjected to the phenotype assay. Table 1. Characterization of monoclonal cells
To generate randomized mutations at rtM204 on the lentipol plasmids, a primer (Rpol204) containing degenerate bases was used to perform FSR. In brief, a DNA fragment was amplified with the primers lenti6744F and Rpol204 (Supplementary Table 1) using pLentipol-D as a template. The plasmid pLentipol-D was constructed previously (Luo et al., 2016), which expresses an HBV (genotype D) polymerase gene. This gel-purified fragment (about 4.5 kb) was used as a mega-primer to substitute the corresponding region on pLentipol-D. After Dpn I digestion, the FSR product was transformed into competent DH5ɑ cells. Clones containing different mutations at rtM204 were characterized by sequencing. Among the 50 clones selected for sequencing, 30 contained a qualified sequence. All 16 possible types of theoretical mutations were identified, except for the mutation of ACG (Supplementary Table S2). The absence of an ACG mutant might be attributed to the paucity of this form in the primer mix, or to the fact that the quantity of clones picked was not sufficient. Of the identified mutation forms, clone numbers varied from 1 to 5. Since ACG encodes threonine as do ACU, ACA, and ACC, we did not further pursue this mutant.
The bacteria containing the 15 kinds of plasmids were respectively mixed and cultured for plasmid preparation. The plasmid mixture was then used to package the lentiviruses. The lentiviruses were transduced into the 293HBV-pol- cell line, which is a modified HEK293 cell line stably expressing all HBV replication-related elements except for the polymerase (Luo et al., 2016). Blasticidin-resistant monoclones were selected by limited titration. To characterize the mutations integrated into these cells, the fragments were amplified from the chromosome DNA of these cells with the primers F260 and RlentiPCR (Supplementary Table S1). Because one of the two primers targets the sequence on the lentiviral vector backbone, the preexisting integrated copy of HBV DNA in 293HBV-pol- cells would not be amplified. Thirty-seven of the 40 clones assayed contained the polymerase gene. The number of clones with different mutation forms varied from 0 to 7 (Table 1). Three mutation forms (ACA, AGC, and AGT) were not found, and thus the M204S mutant was missing. This is possibly due to low titers of corresponding lentiviruses with rt204S in the lentivirus pool, or because these two lentiviruses were not packaged successfully.
The replication capability of each of the polymerase variants was assayed under 0 μmol/L or 20 μmol/L of LAM using the stable cell lines obtained. The replication capability under 0 μmol/L LAM can reflect the “natural” activity, while that under 20 μmol/L of LAM reflects the "fitness" under drug pressure. Intracellular nucleocapsid-associated HBV DNA levels were measured by real-time PCR, and mitochondria DNA was used as reference (Seeger and Sohn, 2014). The F1927 and R2051 primers were used in real-time PCR (Supplementary Table S1), and the reaction system contained 10 μL FastStart Universal SYBR Green Master (Roche Diagnostics, Mannheim, Germany), 2 μL of DNA, 0.1 μL (10 μmol/L) of each primer, and 7.8 μL of H2O. The amplification conditions were as follows: denaturation at 95 °C for 10 min, followed by 35 cycles of denaturation at 94 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 20 s.
Without LAM in the medium, the wild-type polymerase (rt204M) replicated at the highest level (Figure 1B). The rt204I mutant showed 3.039% of the replicating capacity of the wild type, which was similar to that of the rt204N or rt204K mutant. However, the rt204T and rt204R mutants presented much lower replication capacities (0.102% and 0.005%, respectively), indicating a severe negative impact of these mutations on the function of polymerase. The functional deficiency of these mutants is consistent with the pivotal role of the YMDD motif in the activity of polymerase, and explains why these mutations are seldom found in clinical samples (Chayama et al., 1998).Ono-Nita et al. (1999) also reported that the rtM204T, rtM204R, and rtM204K mutants were incompetent for virus replication during transient transfection in vitro.
In the presence of 20 μmol/L LAM, the replication level of the wild-type polymerase was reduced by 6.842 ± 0.983-fold, whereas the polymerases of the rt204I and rt204T mutants remained essentially unchanged (1.201 ± 0.031- and 1.174 ± 0.146-fold reductions, respectively), compared to the observed levels without LAM treatment (Figure 1C). The polymerase of rt204N replicated HBV DNA with a dramatically decreased efficiency (10.723 ± 0.653-fold), indicating a higher sensitivity to LAM, even compared to that of the wild type. The rt204K and rt204R mutants showed moderate sensitivities to LAM, with fold changes of 2.503 ± 0.295 and 2.831 ± 0.225, respectively. All of these mutants showed higher sensitivities than the rt204I mutant, indicating reduced 'fitness' under LAM treatment, which is concordant with the fact that the rt204I variant is predominantly selected over other mutants in patients treated with LAM.
In summary, we developed a novel mutagenesis phenotypic assay with the following characteristics: (1) acquisition of extensive site-specific mutations through a PCR step and FSR; (2) production of a lentivirus pool expressing different polymerase mutants via one-time lentivirus packaging; (3) generation of stable cell lines that replicate HBV DNA by integrated polymerase mutants, which can help to establish an experimental system with lower variation and higher convenience. This randomized phenotypic assay of HBV polymerase provides a useful tool for assessing the replication fitness and drug susceptibility of HBV variants and quasi-species, which may offer new insight for the NUC therapy and the prognosis of patients with chronic hepatitis B.
HTML
-
This work was supported in part by grants from the National Natural Science Foundation of China (81671997), Chongqing Science & Technology Commission (cstc2015jcyjA10023), Chongqing Education Commission (CYB15099), and the Program for Innovation Team of Higher Education in Chongqing (CXTDX201601015). The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
Supplementary figures/tables are available on the websites of Virologica Sinica: www.virosin.org;link.springer.com/journal/12250.
-
Primer 5′–3′ lenti6744F AGGTTTCCCGACTGGAAAGC Rpol204 TTGGCCCCCAATACCACATCATCNNTATAACTGAAAGCCAAACAGTGGG GGA F260 TAGACTCGTGGTGGACTTCTCT RlentiPCR GAATCGAGACCGAGG AGAGGGT F1927 GGAGCTACTGTGGAGTTACTCT R2051 TTGCCTGAGTGCAGTATGGTGA Table S1. Primer sequences
Nucleotides (ANN) Amino acid Number of clones AUG M204 WT 3 AUA M204I 1 AUU M204I 1 AUC M204I 2 ACA M204T 5 ACU M204T 1 ACC M204T 2 ACG M204T 0 AAA M204K 3 AAG M204K 1 AAU M204N 1 AAC M204N 2 AGA M204R 4 AGG M204R 1 AGC M204S 2 AGU M204S 1 Total 30 Table S2. Distribution of mutations among the 30 plasmids sequenced
















 DownLoad:
DownLoad: