HTML
-
Annual influenza epidemics cause approximately 290, 000–650, 000 deaths in addition to causing severe illness in 3–5 million individuals each year (WHO 2018). This threat poses a continuous challenge to the health care and public health system and is an economic burden to individuals and countries. The 1918 H1N1 Spanish flu, which was caused by the most virulent influenza virus to date, infected about 5% of the world's population and had a mortality rate of 2% (Taubenberger and Morens 2006). In recent years, some of the most prevalent highly-pathogenic avian influenza viruses (such as H5N1 and H7N9) have crossed the species barrier, resulting in human infection and death (Liu et al. 2013). Although the pathogenesis of influenza is not fully understood, severe cytokine storm, with markedly higher levels of pro-inflammatory cytokines including interferons (IFNs), tumor necrosis factors (TNFs), interleukins (ILs), and chemokines, has been detected in patients hospitalized with severe influenza infections (Beigel et al. 2005). This aggressive proinflammatory response along with an insufficient antiinflammatory response is commonly referred to as the "cytokine storm" (Liu et al. 2016). Several studies have shown that cytokine storms are directly associated with influenza-induced fatal acute lung injury (ALI) and acute respiratory distress syndrome (ARDS) (Tisoncik et al. 2012; Short et al. 2014). Antiviral treatments, which target viral components, are prone to develop drug-resistant viruses and have a narrow time window for their administration; they are also unable to control the excessive proinflammatory response that occurs in the late stage of infection (Davidson 2018). Therefore, the attenuation of the host inflammatory response with immunomodulators may represent a good strategy for combating severe influenza. In addition, therapeutics designed to target host components are less likely to develop drug-resistant virus strains. In recent years, a few immunomodulatory agents, including proteinase-activated receptor 1 (PAR1) antagonist (Khoufache et al. 2013), sphingosine-1-phosphate (S1P1) receptor 1 agonists (Teijaro et al. 2011), and tyrosine protein kinase (TPK) inhibitors (Florence et al. 2018), have shown protective effects in animal models including alleviating the pulmonary inflammatory reaction and increasing survival rates. However, the lack of efficient cell-based screening models is a great impediment in the development of anti-inflammatory drugs targeting influenza-induced cytokine storms (Davidson 2018). Animal models, despite being ideal for studying potential antiinflammatory therapeutics, are very costly and time-consuming. Therefore, there is an urgent need to establish fast, economical and time-saving screening methods using cellbased models that can be used to screen and develop more potent immunomodulatory agents.
Upon infection of host cells by an influenza virus, they can be subsequently recognized by pattern recognition receptors, such as TLR3/7 and RIG-I, which activate downstream pathways, including the NF-κB pathway and the IRF pathways, leading to the expression of multiple cytokines (Pulendran and Maddur 2015). Among the influenza-induced pro-inflammatory cytokines, some common and important pro-inflammatory factors including IFN-α/β, TNF-α, CCL2, CCL5, CXCL10, IL-1β, IL-6, and IL-8, have been detected in patients with severe influenza (Ramos and Fernandez-Sesma 2015). Based on their roles in influenza mediated inflammatory injuries in the lungs, IL-6, IL-8, TNF-α, CCL2, CCL3, and CCL5 are co-regulated by the NF-κB pathway, and the interferons are regulated by the IRF pathways, which can induce the expression of a panel of ISGs, e.g., CXCL9 and CXCL10, primarily through the STAT pathways and the NF-κB pathway (Iwasaki and Pillai 2014). Although the functions of these cytokines have not been fully elucidated, they can be further categorized into two classes according to their therapeutic effects in animal experiments when being targeted: class one includes factors that do not improve or worsen the disease after gene knockout or antibody neutralization, e.g., CCL5 (Wareing et al. 2004), IFN-β (Mordstein et al. 2008), and IL-1β (Kozak et al. 1995); class two includes factors that, along with gene knockout or antibody neutralization, can significantly increase the rate of survival and reduce inflammation in the lungs, e.g. CCL2 (Damjanovic et al. 2011; Hrincius et al. 2015), and CXCL10 (Ichikawa et al. 2013; Wang et al. 2013). Therefore, the second class of factors appears to be ideal therapeutic targets for influenza-induced inflammation.
In this study, our goal was to establish a reliable cellbased model for high-throughput anti-inflammatory drug screening, along with an efficacy study specific for inhibitors of influenza-induced cytokine release. First, seven human cell lines that are reported to be involved in influenza-induced inflammation were screened to identify an appropriate cell line that can support influenza virus infection and the production of pro-inflammatory cytokines. Using the protein levels of the two important chemokines CCL2 and CXCL10 as readouts, the human monocytic cell line U937 was identified as a suitable cell line and selected to establish a cell-based model. The U937 cell model was validated by testing a panel of agents with reported anti-inflammatory or antiviral activities in vivo and a drug library comprised of 1280 compounds, most of which are FDA-approved drugs. Our results indicate that the human U937 cell line can be used as a model to study the influenza-induced cytokine release and in highthroughput screening of drugs that target components of the influenza-induced cytokine storm.
-
The Madin-Darby Canine Kidney cell line MDCK (CCL- 34), human lung adenocarcinoma cell line A549 (CCL- 185), human monocyte cell lines U973 (CRL-1593.2) and THP-1 (TIB-202), and human promyeloblast cell line HL- 60 (CCL-240) were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA). MDCK cells were cultured in Dulbecco's modified Eagle's medium (DMEM), and the other cells were maintained in RPMI 1640 medium. Both the DMEM and RPMI 1640 medium were supplemented with 10% fetal bovine serum (FBS, Gibico) and 1% penicillin–streptomycin. The differentiation of U937 and THP-1 cells into cells possessing a macrophage-like phenotype was achieved by induction with 100 ng/mL of phorbol-12-myristate-13-acetate (PMA; Sigma-Aldrich) for 24 h (Garcia et al. 1999; Genin et al. 2015). To induce HL-60 cells to differentiate into neutrophil-like cells, HL-60 cells were cultured in growth media (RPMI1640 + 10% FBS) supplemented with 1.25% dimethyl sulfoxide (DMSO) for 5 days, as described previously (Millius and Weiner 2009). All cells were cultured at 37 ℃ in an incubator with 95% humidity and 5% CO2.
The strains of influenza viruses used in this study, including A/PuertoRico/8/1934 (H1N1), A/Human/Hubei/ 3/2005 (H3N2), A/Duck/Hubei/216/1983 (H7N8), and B/Human/Hubei/1/2007 (IBV), were provided by the virus collection at the Wuhan Institute of Virology, Chinese Academy of Sciences, China. Virus stocks were prepared in 10-day-old embryonated chicken eggs. The virus titers were determined using the 50% tissue culture infective dose (TCID50) assay in MDCK cells using the method developed by Reed and Muench (1938).
-
A library of compounds used for drug screening was purchased from MicroSource Discovery Systems, Inc. (Gaylordsville, CT, USA). The library consists of 1280 compounds with broad biological activities and structural diversity and was assigned the name FDA-drug library. This library includes: (1) more than 1000 drugs that have been approved by the FDA or have reached clinical trials in the United States; (2) drugs of international collections that are in clinical use in Europe and/or Asia, but have not yet received FDA approval; (3) a unique collection of approximately 100 pure and natural products and their derivatives. These compounds all have known biological activities with a purity of > 95% and are provided in a DMSO stock solution at a concentration of 10 mmol/L.
Compounds used in the validation of the U937 cell model, including the tyrosine protein kinase (TPK) inhibitor ibrutinib (S2680), protease-activated receptor 1 (PAR1) antagonist vorapaxar (S8067), sphingosine-1- phosphate receptor 1 (S1PR1) agonists ozanimod (S7952), macrolide antibiotics ascomycin (S7411), non-selective cyclooxygenase (COX) inhibitor ibuprofen (S1638), COX2 selective inhibitor etoricoxib (S4651), and peroxisome proliferator-activated receptor (PPAR)-γ agonist rosiglitazone (S2505), were purchased from Selleck Chemicals (Shanghai, China). Ribavirin was purchased from the Sigma Chemical Company (Sigma-Aldrich, MO, USA). Oseltamivir (GS 4071) was purchased from Toronto Research Chemicals (Toronto, Canada). All compounds were initially dissolved in DMSO (Sigma-Aldrich, MO, USA).
-
2'-(4-Methylumbelliferyl)-α-D-N-acetylneuraminic acid (MUNANA, Sigma, M8639) is a fluorescent substrate for the neuraminidase (NA) of influenza viruses and is used to detect the replication levels of influenza viruses; therefore, it can also be used to measure the inhibitory effect of agents against influenza virus infection (An et al. 2014). Briefly, the virus-containing culture supernatant was transferred to a black opaque 96 or 384 well plate (PerkinElmer, 6005270 or 6007270) and mixed with 20 μmol/L of MUNANA dissolved in MES solution (33 mmol/L 2-[N-morpholino] ethanesulfonic acid and 4 mmol/L CaCl2, pH = 6.5), followed by incubation at 37 ℃ for 1 h. The reaction was terminated by the addition of stop solution (0.14 mol/L NaOH in 83% ethanol). Fluorescence intensity was measured at an excitation wavelength of 355 nm and an emission wavelength of 485 nm using multi-label plate readers (Envision2103, PerkinElmer, USA).
-
PerkinElmer's bead-based AlphaLISA immunoassays are designed for the detection of analytes in biological samples. The AlphaLISA assay was used to quantify the protein levels of cytokines and chemokines. AlphaLISA detection kits used include CCL2 (PerkinElmer, AL244), CXCL10 (PerkinElmer AL259), IL-6 (PerkinElmer AL223), and IL-8 (PerkinElmer AL224). All AlphaLISA assays were performed according to the manufacturer's protocols. Briefly, The AlphaLISA was performed in white opaque 384 well plates (PerkinElmer, 6007290) using 5 μL of a sample, 25 μL 1 × AlphaLISA buffer (PerkinElmer AL000C) containing 25 μg/mL of specific acceptor beads and 2.5 nmol/L of a specific biotinylated antibody. The plates were incubated at room temperature for 1 h and then 25 μL of streptavidin donor beads (final concentration 80 μg/mL) was added to each well. The plates were incubated at room temperature for 0.5 h in the dark. The plates were read using an EnVision multi-label plate reader (Envision2103, PerkinElmer, USA).
-
We used the CellTiter 96 AQueous One Solution Cell Proliferation kit (Promega, G3580) to measure the viability of the cells. Cell viability is assessed by detecting the capability of the cells to convert 3-(4, 5-Dimethylthiazol-2- yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) to formazan. Specifically, after removing the supernatant (170 μL for 96 well plates and 70 μL for 384 well plates) of the treated U937 cells, a medium (RPMI 1640 + 10% FBS) containing 20% of the detection reagent was added and incubated at 37 ℃ for 1 h. After centrifugation of the test plate at 500 ×g for 3 min using a swing-out rotor, the optical density (OD) value at the specific wavelength of 490 nm (OD490) and the reference wavelength of 630 nm (OD630) was measured using the EnVision Multilabel Plate Reader. The final absorbance is equal to the value of OD490 minus the value of OD630 in order to rule out the effects of excess cell debris, fingerprints, and other non-specific absorption.
-
In the primary screen, 1280 compounds in the FDA-drug library were dissolved in DMSO at a concentration of 10 mmol/L and added to four 384-well source plates (Labcyte, LP-0200) with 320 compounds per plate. Subsequently, 320 nL of each compound, positive control drugs, or DMSO were transferred to four sterile, clearbottom view 384-well plates (PerkinElmer, 6007460) using an acoustic droplet ejection (ADE) system (Echo 550, Labcyte, CA, USA). Forty μL of complete medium (RPMI 1640 + 10% FBS + 1% penicillin–streptomycin) was added to each well to dilute the drug to a final concentration of 40 μmol/L. To prepare for viral infection, U937 cells were resuspended at a density of 1 × 106 cells/mL in complete medium and infected with 0.05 multiplicity of infection (MOI) of the A/PuertoRico/8/1934 (H1N1) virus. Immediately after mixing, 40 μL of the cell-virus mixture was added to each well in the compound-containing 384-well plates with a cell density of 40, 000 cells/well; 40 μL of uninfected cells were also added to the negative control wells. After incubating at 37 ℃/5% CO2/95% relative humidity for 48 h, the cell culture plates were centrifuged at 500 ×g for 3 min using a swing-out rotor, and 70 μL of supernatant per well were taken for the detection of NA activity and cytokine levels. The remaining cells were used to test for cell viability.
In the confirmation screen, the serially diluted hit compounds (0.04–90 μmol/L) were added to 384-well plates. The U937 cells and influenza virus were added and incubated for 48 h in the presence or absence of the drugs as described previously to confirm the inhibitory effect of the drug and to study the kinetics of the drug response. In parallel, the cytotoxicities of hit compounds were determined in the same conditions but without viral infection. Finally, the half maximal inhibitory concentration (IC50), half maximal toxicity concentration (CC50) and selective index (SI) of the hit compounds were determined.
-
Viral titers were determined using a TCID50 assay. Briefly, MDCK cells were seeded into 96-well cell culture plates at a density of 20, 000 per well. 24 h later, tenfold serial dilutions of the virus solution were inoculated on an MDCK monolayer at 37 ℃ for 72 h, and the cytopathic effects (CPE) were examined. The virus titers were calculated using the method developed by Reed and Muench (Reed and Muench 1938).
-
U937 cells were collected 12 h post-infection with the H1N1 PR8 virus at 0.1 MOI, and the total RNA was extracted according to the protocol in the E.Z.N.A Micro Elute Total RNA kit (OMEGA, R6831). Reverse transcription was performed using the M-MLV Reverse Transcriptase (Promega, M170A) and random hexamer primer. Quantitative mRNA expression analysis was performed using the SYBR green method (iTaqTM Universal SYBR® Green Supermix, Bio-Rad, 1725121) according to the manufacturer's instructions. The PCR Conditions were as follows: denaturing at 95 ℃ for 30 s; 40 cycles of 94 ℃ for 5 s and 60 ℃ for 30 s. To avoid intra-plate errors, all experiments were performed in a 96-well plate in triplicate per sample. Primer sequences of the cytokines and chemokines used in the experiments were all taken from published literature (Giulietti et al. 2001).
-
The IC50 and CC50 of the compounds were determined by non-linear regression using the Graph Pad Prism 5.0 software. The data in the figures are presented as the mean ± SEM. The Z' value is statistically calculated using a previously reported method (Zhang et al. 1999). Specifically, the Z' value is calculated as follows: Z' value = 1 - [(3 × SD of sample + 3 × SD of control)/|average of sample - average of control|], where the SD value represents the standard deviation of the fluorescence signal.
Cell Lines and Virus Strains
Chemicals
Neuraminidase Activity Assay
AlphaLISA Assay
Cell Viability Assay
High-Throughput Screening (HTS) of FDA-Drug Library
Measurement of Virus Titer
Total RNA Extraction and Real-Time PCR
Statistical Analysis
-
Lung epithelial cells, neutrophils, and monocytes/macrophages all play important roles in the cytokine storm induced by influenza virus infection and the subsequent effects of ALI and ARDS (Short et al. 2014). In order to identify a human cell line that can support influenza virus infection and the production of important proinflammatory cytokines, while also being suitable for screening anti-influenza drugs suppressing cytokine release, we examined seven human cell lines, including the epithelial lung A549 cells, promyelocytic HL-60 cells, neutrophil-like iHL-60 cells, monocytic TPH-1 cells, macrophage-like iTPH-1 cells, monocytic U937 cells, and macrophage-like iU937 cells, for their abilities in supporting influenza virus infection and the production of proinflammatory cytokines based on signal-to-background ratios (SBRs) during virus replication and CCL2/CXCL10/ IL-8/IL-6 expression levels (Table 1). Among the cell lines tested, A549 has lower SBRs in viral replication and proinflammatory cytokine secretion when infected with H1N1 PR8 influenza virus at MOI of 0.1 and cultured for 48 h, while HL-60 and iHL-60 can support influenza virus replication under the same conditions, but they only induce specific cytokine expression (CCL2 for HL-60, IL-6 for iHL-60). Therefore, these cell lines are not suitable for developing cell models for screening of anti-influenza drugs suppressing cytokine release. The human monocytic U937 cell line was found to support efficient replication of influenza virus H1N1 PR8 strain and the production of the four pro-inflammatory cytokines tested; additionally, it also had the highest SBRs for both viral replication and proinflammatory cytokine expression. Interestingly, THP-1 cell line, which is also monocytic cell, was shown to support influenza virus replication; however, it only expressed two of the four pro-inflammatory cytokines with very low expression levels under the same conditions. It is worth noting that the U937 and THP-1 cells were less supportive of influenza virus infection after being induced into macrophage-like cells and the SBRs of viral replication decreased from 7.3 and 5.8 to 2.4 and 1.5, respectively. Although the total expression levels of inflammatory cytokines in iU937 and iTHP-1 were significantly increased (data not shown), the amount of cytokine expression in the uninfected group was also greatly increased, which resulted in the lower SBRs of the three pro-inflammatory cytokines. These results indicate that the un-induced monocyte cell line U937 has the greatest potential to be a cell model for screening anti-inflammatory agents among the seven cell types tested.
Name Cell-type Signal to background ratio (SBR)a NAb CCL2c CXCL10c IL-8c IL-6c A549 Lung epithelial cell line 2.3±0.5 1.6±0.1 1.5±0.3 1.9±0.1 3.6±0.1 THP-1 Monocyte cell line 5.8±2.1 3.2±0.7 1.7±0.2 2.0±0.2 1.2±0.1 iTHP-1d Macrophage-like cell 1.5±0.6 1.4±0.2 3.4±0.2 1.2±0.1 1.1±0.1 U937 Monocyte cell line 7.3±1.1 16.2±2.5 23.6±3.3 17.9±2.3 13±3.7 iU937d Macrophage-like cell 2.4±0.7 1.1±0.1 2.5±0.2 1.2±0.2 1.6±0.2 HL-60 Promyeloblast 6.1±0.6 13.0±2.7 1.2±0.2 4.4±0.1 1.3±0.3 iHL-60e Neutrophil-like cell 4.1±0.4 1.4±0.3 1.0±0.1 2.2±0.3 13.2±2.6 aCells were incubated with 0.1 MOI of A/PuertoRico/8/1934 (H1N1) influenza virus and tested for the signal to background ratios (SRBs) at 48 h post infection. The SBR was calculated from the fluorescence value of the infected wells (signal) divided by the uninfected control wells (background) for each assay. b NA (neuraminidase), measured by neuraminidase activity assay, represents the replication level of the influenza virus. cCytokine expression was measured by AlphaLISA. diU937 and iTHP-1 cells were prepared from U937 and THP-1 cells induced by 100 ng/mL TPA for 24 h. eiHL-60 was prepared from HL-60 induced by 1.25% DMSO for 5 days. Table 1. Identification of U937 cell line as a cell-based model for drug discovery against cytokines induced by influenza virus infection.
-
To confirm whether the U937 cell line is capable of supporting influenza virus replication and the secretion of proinflammatory cytokines, they were infected with the influenza virus PR8 strain at different MOIs. Viral replication levels were determined according to neuraminidase (NA) levels and viral titers in a time course assay. As shown in Fig. 1A, NA activities in the supernatants increased with increasing MOIs of the viruses and reached a peak level at 48 hpi (hours post infection). Interestingly, both the culture time and the MOI can affect the viral titers of the supernatants of infected U937 cells (Fig. 1B). We found that the viral growth curves in U937 cells determined by the TCID50 endpoint dilution assay were consistent with what was determined using the NA activities when the MOIs were equal to or less than 0.1, suggesting that under such infection MOI conditions, the NA activities correlate well with the influenza virus titers. The TCID50 growth curves at high MOIs are partially inconsistent with the NA growth curves, in that the TCID50s reached peak levels at 24 hpi, possibly due to the cell death caused by virusinduced cytopathic effects with longer culture times (Fig. 1A–1C). Next, we tested the mRNA levels of the seven major pro-inflammatory cytokines induced postinfection in the influenza-infected U937 cells. As shown in Fig. 1D, at 12 hpi, the mRNA levels of CCL2, CCL5, CXCL10, IL-6, IL-8, IFN-β and TNF-α in U937 cells increased by 26.3, 1.7, 318.3, 27.5, 7.5, 344.2 and 2.15 times, respectively, indicating that PR8 infection can stimulate the transcription of pro-inflammatory cytokines in U937 cells. To study the correlation between the production of cytokines and virus MOIs, we then measured the protein levels of CCL2 and CXCL10 in the supernatants of the PR8 virus infected U937 cells. As shown in Fig. 1E and 1F, there was no apparent production of cytokines observed at 12 hpi, and the protein levels of CCL2 and CXCL10 were found to be at low levels at 24 hpi, reaching their highest levels at 48 hpi. It is worth noting that the levels of both CCL2 and CXCL10 increased with decreasing MOIs and reached their highest levels when the MOIs were around 0.025–0.05. However, as the MOI decreased below that range, i.e. 0.0125, the productions of the pro-inflammatory cytokines CCL2 and CXCL10 were observed to greatly decrease. These results indicate that U937 cells can support the replication of the influenza virus PR8 as well as the production of multiple cytokines. More importantly, infection of U937 cells with a relatively low MOI is critical for the production of the influenza-induced cytokine storm; infection with a high MOI can cause severe CPE and a low MOI can result in a low level of viral replication, both conditions can attenuate influenza-induced cytokine expression.
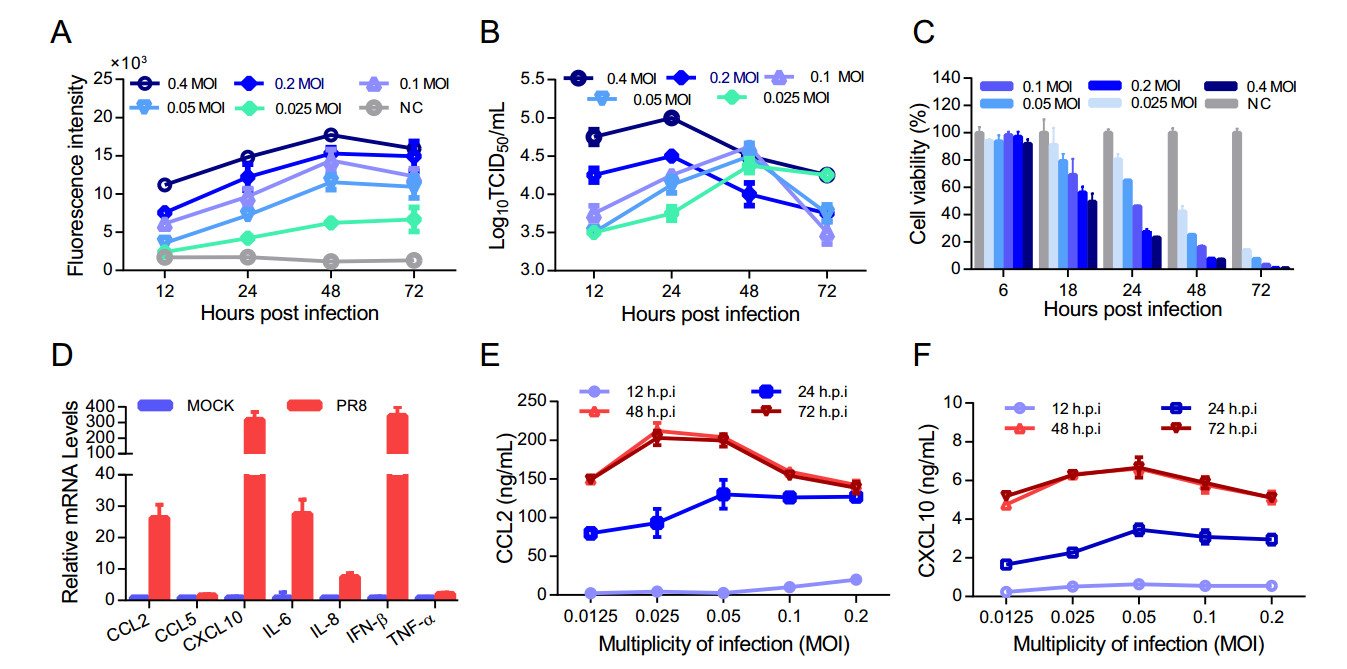
Figure 1. U937 cells support H1N1 virus replication and express multiple cytokines. U937 cells were infected with the PR8 (H1N1) virus at different MOIs and incubated at 37 ℃ for 1 h. The uninfected virus was washed away through centrifugation and cells were cultured at 37 ℃ for 72 h with complete medium (RPMI1640 + 10% FBS). Supernatants were collected at the indicated time points for the detection of NA activities (A) and virus titers (B). The cell viabilities were determined using the MTS method (C). The mRNA levels of 7 important pro-inflammatory cytokines in the U973 cells were determined by quantitative real-time PCR analysis at 12 h post infection (D). The protein levels of CCL2 (E) and CXCL10 (F) in the supernatants of U937 cells infected with the PR8 virus at different MOIs were determined using AlphaLISA. Each data point in the figure was repeated three times and the values were presented as mean ± SEM. NC represents cell control.
-
Influenza viruses are classified into types A, B, C and D and make up four of the seven genera of orthomyxoviridae. The type A viruses are the most virulent pathogens and can also be subdivided into different serotypes based on the antibody response to the viral components HA and NA. Influenza B viruses almost exclusively infect humans and can cause severe disease; they have only one serotype. Each of these influenza viruses may replicate and induce a cytokine storm in host cells differently (Hay et al. 2001). To investigate whether U937 cells can support the replication of different subtypes of influenza viruses and as well as the expression of the major pro-inflammatory cytokines in addition to the IAV H1N1 strain we initially used, three other subtypes of influenza virus strains from type A and one strain from type B, including H3N2, H7N8, and IBV, were selected and used to infect U937 cells. All four virus strains were able to replicate in U937 cells, although IBV was shown to replicate much less efficiently, as shown in Fig. 2A. Next, we examined the expression of CCL2, CXCL10, and IL-8 in the supernatants of the infected U937 cells. H1N1, H3N2, and H7N8 virus strains were shown to induce the expression of CCL2, CXCL10, and IL-8 (Fig. 2B–2D). Interestingly, IBV, which was shown to replicate inefficiently in U937 cells, activates the expression of the three cytokines as efficiently as the other three IAV strains (Fig. 2). These results indicate that U937 cells can support the replication of different types and subtypes of influenza viruses and induce the expression of multiple pro-inflammatory cytokines in response to these virus infections. In addition, the infection of a relatively low MOI of influenza virus in U937 cells is critical and sufficient for the production of multiple cytokines.
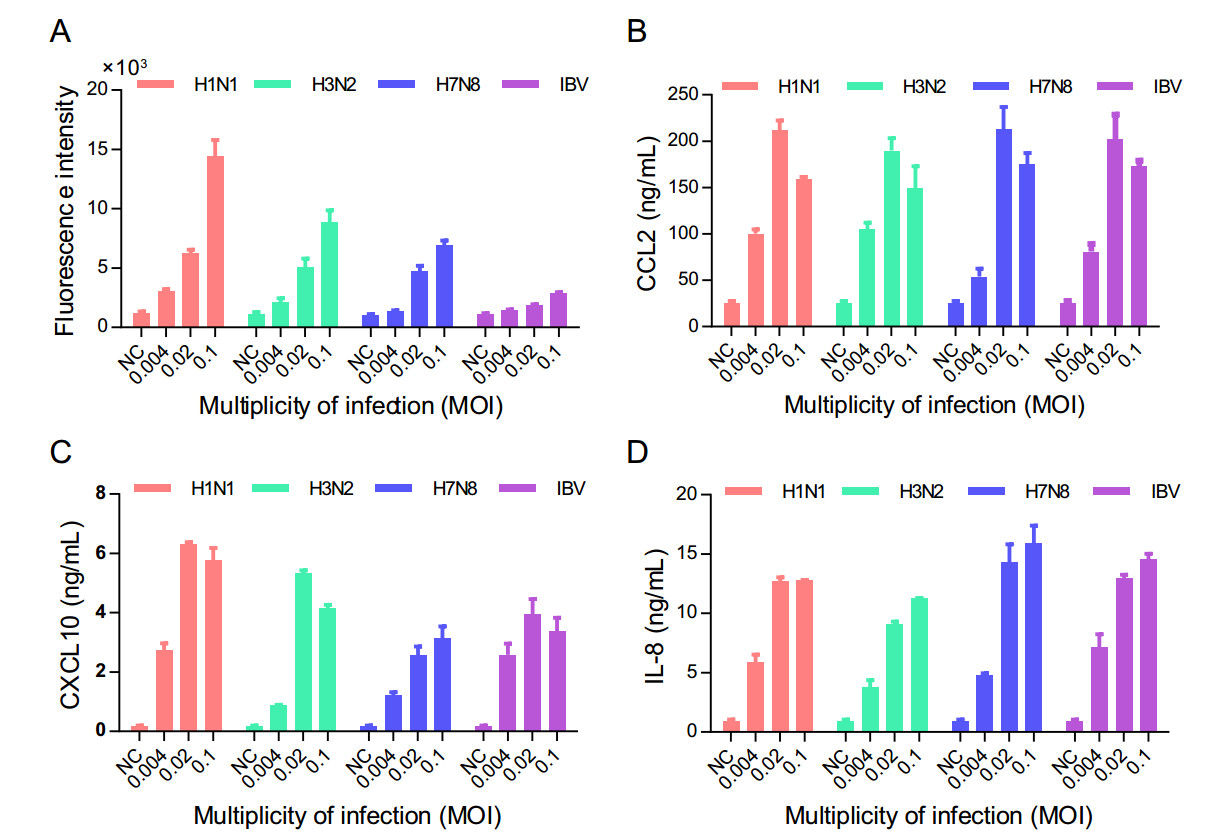
Figure 2. U937 cells support the replication of multiple subtypes of influenza viruses and the expression of pro-inflammatory cytokines. Four subtypes of influenza viruses, including A/PuertoRico/8/1934 (H1N1), A/Human/Hubei/3/2005 (H3N2), A/Duck/Hubei/216/1983 (H7N8), and B/Human/Hubei/1/2007 (IBV), were used to infect U937 cells at different MOIs at 37 ℃ for 1 h, and the uninfected viruses were washed away using centrifugation. The cells were cultured at 37 ℃ for 48 h with complete medium (RPMI1640+ 10% FBS). The supernatants were collected and used to detect the levels of NA activity (A), and the protein levels of CCL2 (B), CXCL10 (C) and IL- 8 (D) using a neuraminidase assay and AlphaLISA, respectively. Each data point in the figure was repeated three times and the values were presented as mean ± SEM. NC represents cell control.
-
To validate the U937 cell model, we tested a panel of immunomodulatory and antiviral agents with known in vivo activities (Table 2). These immunomodulatory agents, including the TPK inhibitor ibrutinib, PAR1 antagonist vorapaxar, S1PR1 agonist ozanimod, PPARγ agonist rosiglitazone, and macrolides antibiotics have been reported to improve the survival rate and pathology in influenza-infected mice. As expected, most of these immunomodulatory agents, with the exception of rosiglitazone, showed anti-inflammatory activities in the U937 cell model, as determined by their inhibition of CCL2 and CXCL10 expression. The antiviral agents, including the macrolide antibiotic ascomycin and the antiviral drugs ribavirin and Oseltamivir, were shown to have antiviral effects in the U937 cell model, as determined by the neuraminidase activity assay (Table 2). In contrast, some of the anti-inflammatory agents, including the COX non-selective inhibitor ibuprofen and the COX2-specific inhibitor etoricoxib, which have been reported to have no improvement on the survival rate of influenza-infected mice when used alone, were shown to have no inhibitory effect on the production of CCL2 and CXCL10 in U937 cell model (Table 2). Though the diversity and numbers of agents tested here were limited, these results suggest that, when using NA as an antiviral readout and CCL2/CXCL10 expression as anti-inflammatory readouts, the U937 cell model can be used to screen antiviral and immunomodulatory agents against influenza infection.
Chemical
nameCC50a IC50b
NASId
NAIC50c
CCL2IC50c
CXCL10SId
CCL2SId
CXCL10Target and possible mechanisme In vivo f Referencesg Ibrutinib 26.5 –h – 0.62 0.96 42.7 27.6 TPK inhibitor +i Florence et al.(2018) Vorapaxar > 80 – – 5.2 13.3 > 15.4 > 6 PAR1 antagonist + Khoufache et al.(2013) Ozanimod 26 – – 7.7 10.7 3.4 2.4 S1PR1 agonist + Teijaro et al. (2011) Ascomycin 62 21.3 2.9 8.1 9.9 7.7 6.3 Macrolides antibiotics + Sato et al. (1998) Ribavirin > 160 26.2 > 6.1 – – – – Anti-influnenza + Smee et al. (2008) Oseltamivir > 100 0.58 > 172.4 – – – – Anti-influnenza + Tsai et al. (2015) Rosiglitazone > 100 – – – – – – PPARγ agonist + Moseley et al.(2010) Etoricoxib > 50 – – – – – – COX-2 inhibitor ±j Zheng et al. (2008) Ibuprofen > 50 – – – – – – COX non-selective inhibitor ± Lauder et al. (2011) aCC50: 50% cytotoxic concentration (μmol/L) determined by MTS assay. bNA IC50: 50% inhibition concentration (μmol/L) determined by NA activity assay. cCCL2/CXCL10 IC50: 50% inhibition concentration (μmol/L) determined by AlphaLISA. dSI (Selective Index): a ratio of CC50/ IC50. eTarget and possible mechanism attributed to the compound as stated on either its "label" or literature. fIn vivo: the protective effect of mice from lethal influenza infection. gRef.: the reference of the reported protection effect of antiviral or immunomodulation agents in vivo. h"–": lower than 50% inhibition or not effective. i"+": Agent used alone can protect mice against lethal influenza virus infection according to the literature. j"±": Agent is effective in combination therapy, but when used alone cannot protect mice against lethal influenza virus infection. Table 2. Validation of U937 cell model with a panel of antiviral and immunomodulatory agents.
-
After the optimization of several parameters, including the number of cells per well, the MOI, and culture time, a U937 cell-based HTS model was established. Briefly, U937 cells were cultured in 384-well plates at a density of 1 × 106/mL, followed by infection with 0.05 MOI of influenza virus. The plates were cultured for 48 h at 37 ℃. Viral replication and the expression levels of cytokines were determined using a neuraminidase assay and AlphaLISA, respectively. The performance of our U937 cellbased HTS platform was evaluated by screening the FDAdrug library to identify agents that can inhibit the induction of the expression of CCL2 and CXCL10 by influenza infection (Fig. 3A). At a concentration of 20 μmol/L, 1280 compounds were screened on four 384-well plates. The SBRs of NA in the four plates were between 7.5 and 15, as shown in Fig. 3B. The SBRs of CCL2 and CXCL10 were between 17.6 and 21.5, and 20.8 and 28.3, respectively. The average Z' factor for NA, CCL2, and CXCL10 were 0.74, 0.75 and 0.84, respectively; all of the Z' factors were greater than 0.65, suggesting that the U937 cell model is robust and desirable for the HTS of immunomodulatory agents against the production of pro-inflammatory cytokines induced by influenza virus infection.
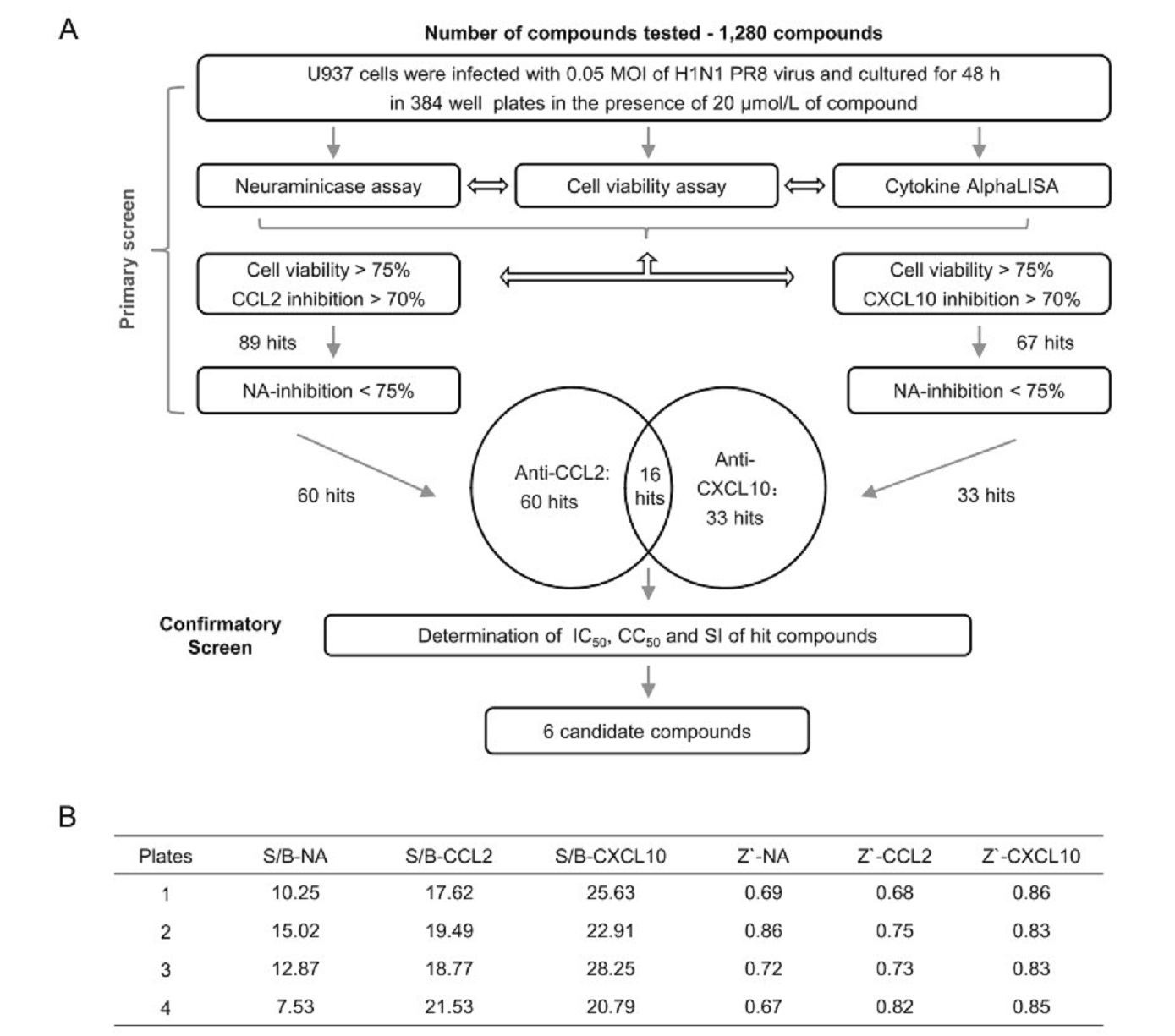
Figure 3. Establishment and implementation of a quantitative HTS using the U937 cell model. A Flow chart for the HTS of the 1280 drug library for immunomodulatory agents against influenza infection. The flow chart includes the steps and conditions for the entire screening process and also lists the selection criteria of the hit compounds and the number of hits obtained after each screening. The Venn diagram in the flow represents the cross-check of anti-CCL2 hits and antiCXCL10 hits and the results. B Fluorescence values representing NA activity, CCL2 and CXCL10 levels from infected and non-infected cells from all four 384-well plates were used to calculate the signalto-background ratio and Z'-factor scores. The calculation method used for the Z' factor is described in the statistical analysis section in materials and methods.
In the screen of the FDA-approved drug library, a hit was defined as a compound with cell viability > 75%, and an inhibition rate for CCL2 or CXCL10 > 70%, and an NA inhibition rate < 75%. We identified 60 hits with an anti-CCL2 effect with a hit rate of 4.7%, and 33 hits with an anti-CXCL10 effect with a hit rate of 2.6%. Interestingly, when analyzing the composition of the hit compounds with anti-CCL2 and anti-CXCL10 effects, listed in Supplementary Table S1, we found that most of the traditional anti-inflammatory drugs, such as steroidal anti-inflammatory drugs, cox inhibitors, prostaglandin synthetase inhibitor, histamine antagonist, 5-HT2 receptor antagonist, b-adrenergic blocker, and cholinergic receptor inhibitor, were identified as hits with anti-CCL2 effects, but not as hits with anti-CXCL10 effects. In addition to traditional antiinflammatory drugs, many agents with new targets were identified as well, which include the majority of the 16 hits found in both the anti-CCL2 and anti-CXCL10 hits, including HMG-CoA reductase inhibitors, DNA gyrase/topoisomerase IV inhibitors, ion channel inhibitors, and tyrosine kinase inhibitors. It appears that the target distributions of CCL2 and CXCL10 are different but related to some extent, suggesting that they are co-regulated by certain signaling pathways, and specifically regulated by their own signaling pathways.
-
In the primary single dose (20 μmol/L) screen using the influenza virus infected U937 cell model on 384-well plates, we identified 16 hit compounds with an anti-inflammatory effect (inhibiting both CCL2 and CXCL10) without an antiviral effect. To further confirm and evaluate their efficacies in repressing cytokine expression, we tested all 16 hit compounds for their kinetic inhibition of CCL2 and CXCL10 and cell viabilities in U937 cells. Their IC50s and CC50s were determined. Compounds with an IC50 > 10 μmol/L or shown to have no effect were considered to be inactive, 10 out of 16 hit compounds met this criterion (data not shown). Compounds with an IC50 < or = 10 μmol/L were considered to be active; 6 out of the 16 primary hit compounds met this criterion, they are dasatinib, pitavastatin, simvastatin, fluvastatin, protriptyline, and levofloxacin (Fig. 4A–4F). The cytotoxicity of the six hit compounds on uninfected U937 cells was determined, as shown in Fig. 4 (right panel). The CC50s were all greater than 90 μmol/L, except for simvastatin which had a CC50 of 66.8 μmol/L. To exclude compounds with antiviral activities that may lead to a reduction in CCL2 and CXCL10 levels, we examined the antiviral effects of the six hit compounds in the same confirmation experiments simultaneously (Fig. 4 right panels). Surprisingly, all 6 of the hits were shown to promote viral amplification to varying degrees, and viral replication was shown to increase with increasing drug concentrations, suggesting that these compounds may also inhibit the expression or functions of certain host factors associated with the innate antiviral response that correlated to the influenza-induced cytokine storm. Based on the kinetic inhibition of the two important cytokines CCL2 and CXCL10, and the IC50s and SIs (Fig. 4 and Table 3), we identified 6 compounds with potent inhibitory effects on influenza-induced expression of cytokines in U937 cells. Taken together, our results suggest that the U937 cell model is suitable for high throughput screening of antiinflammatory drugs against influenza-induced cytokine release.
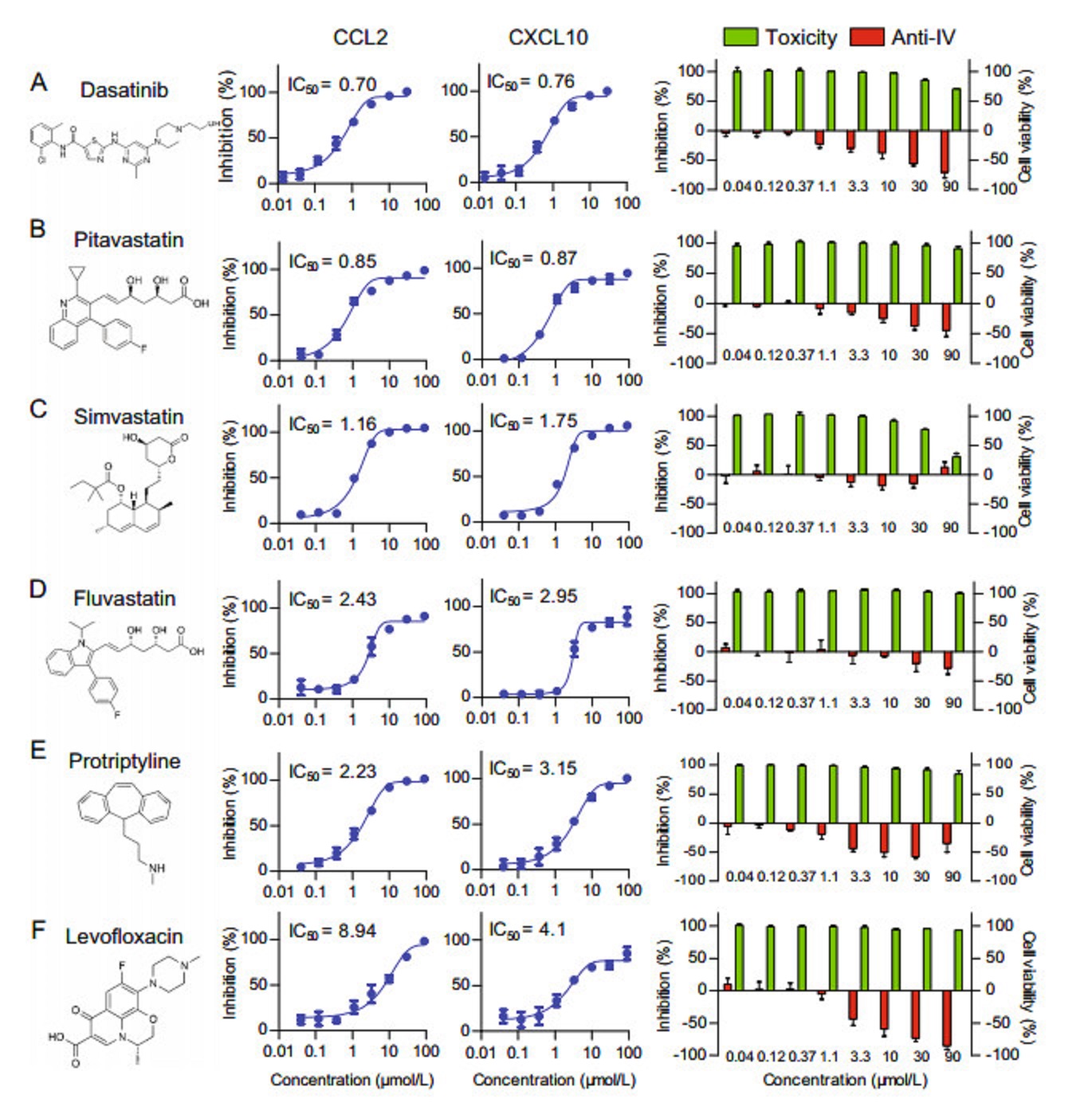
Figure 4. Six hit compounds were confirmed to inhibit the production of CCL2 and CXCL10 in the U937 cell model. The kinetics of the inhibition of CCL2 and CXCL10 expression of the primary hit compounds was tested in the U937 cell model for Dasatinib (A), Pitavastatin (B), Simvastatin (C), Fluvastatin (D), Protriptyline (E), Levofloxacin (F). The structures of the six compounds are shown in the left panel. The inhibitory effects on CCL2 and CXCL10 are shown in the middle panels. The compound toxicity (green) and antiviral activity (red) expressed as a histogram are shown in the right panel. Each data point represents the mean ± SEM. The IC50 values for inflammatory factors are listed in the figure and were calculated using Graph Pad Prism 5.0 software.
Chemical name IC50
CCL2IC50
CXCL10CC50 SI
CCL2SI
CXCL10Approved of intended use Possible mechanism and target Dasatinib 0.63 0.71 > 90 > 142.9 > 126.8 Antineoplastic Tyrosine-protein kinases inhibitor Pitavastatin 0.85 0.87 > 90 > 105.9 > 103.4 Antilipemic HMG-CoA reductase inhibitor Simvastatin 1.16 1.75 66.8 57.6 38.2 Antilipemic HMG-CoA reductase inhibitor Fluvastatin 2.43 2.95 > 90 > 37.0 > 30.5 Antilipemic HMG-CoA reductase inhibitor Protriptyline 2.23 3.15 > 90 > 40.4 > 28.6 Antidepressant Serotonin reuptake inhibitor Levofloxacin 8.94 3.12 > 90 > 10.1 > 28.8 Antibiotic DNA gyrase and topoisomerase IV inhibitor Table 3. Six compounds were confirmed to inhibit influenza-induced CCL2/CXCL10 expression in U937 cell model.
Identification of the U937 Cell Line as a Cell-Based Model for Drug Discovery Against Cytokines Induced by Influenza Virus Infection
U937 Cells Support the Replication of Influenza Virus H1N1 and Produce Multiple Cytokines upon Influenza Virus Infection
U937 Cells Support the Replication of Multiple Subtypes of Influenza Viruses and the Expression of Pro-Inflammatory Cytokines
The U937 Cell-Based Model was Validated Using a Panel of Immunomodulatory and Antiviral Agents with Known Activities In Vivo
U937 Cell-Based HTS Model was Established and Validated by Screening an FDA-Drug Library
Six Hit Compounds were Confirmed to Inhibit the Production of Multiple Pro-Inflammatory Cytokines on U937 Cell Model
-
Due to the narrow therapeutic window for the administration of anti-flu drugs, typically within 2 days of the onset of symptoms, and the association of excessive inflammatory responses with severe influenza pneumonia, the treatment of influenza pneumonia cannot rely solely on antiviral drugs. Combination therapies that include both antiviral agents and anti-inflammatory agents represent a potential solution in curing influenza pneumonia. Immunomodulators that can inhibit the excessive inflammatory response in the lungs of patients infected by influenza viruses are urgently needed. However, the use of traditional anti-inflammatory drugs, including corticosteroids, during an influenza infection has been found to have either no effect or even detrimental effects in clinical retrospective studies (Hui and Lee 2013; Ramos and Fernandez-Sesma 2015). Therefore, the use of selective immunomodulatory agents to specifically inhibit the excessive expression of pro-inflammatory cytokines in influenza pneumonia is a potentially valid strategy. However, currently, it is rare to find influenza-related anti-inflammatory drugs either in clinical trials or in preclinical research. Several animal models for influenza infection, including mouse, ferret, and guinea pig models, are suitable for the evaluation of potential anti-inflammatory drugs, however, they are not suitable for HTS screening of compound libraries comprised of synthetic small molecules or natural products. There are a few existing influenza infection cell models suitable for high-throughput antiviral drug screening, those include kidney cell models (MDBK or MDCK) (Gerritz et al. 2011; An et al. 2014), human epithelial cell model (A549) (Hoffmann et al. 2008). However, the current cell models are not suitable for screening of immunomodulating agents for influenza treatment. Therefore, the establishment of a cell-based influenza virus infection model that can be used for both antiviral and anti-inflammatory drug discovery is very valuable and urgently needed.
Cell models are quite different from animal models in that they do not show indications of viral pneumonia found in the lungs of animals. To establish a useful cell model, the right cell type must be chosen; one that plays an important role in influenza-induced cytokine storms and has the appropriate readouts (e.g. induces the expression of the most important cytokines upon influenza infection). Lung epithelial cells, neutrophils, and monocytes/macrophages all play important roles in the influenza-induced cytokine storm and the subsequent ALI and ARDS (Short et al. 2014). In order to identify a human cell line that can support multi-types and -subtypes of influenza virus infection and the production of important pro-inflammatory cytokines, we examined seven human cell lines, including epithelial cell, neutrophils, and monocytes/macrophages, that are important for the pathogenesis of severe influenza and found that monocytic U937 cells can support the infection and replication of the three IAV and one IBV strains tested. Next, we measured the expression of 7 cytokines, including CCL2, CCL5, CXCL10, IL-6, IL-8, IFN-β, and TNF-α, by quantifying their mRNA levels. A strong response was observed for five of the seven cytokines. For the purpose of using U937 cells as a model for the influenza-induced cytokine production, we measured the correlation between the kinetic expression of CCL2 and CXCL10 at different time points and the MOIs of the viruses used for infection and found that the expression of CCL2 and CXCL10 reached their peak levels when the U937 cells were infected with an influenza virus at an MOI between 0.025 and 0.05. Furthermore, the U937 cell model was demonstrated to support complete viral replication and the expression of CCL2, CXCL10, and IL8 when infected by all of the four subtypes of influenza viruses tested. Therefore, the U937 cell model exhibits great potential for drug discovery of anti-inflammatory and antiviral agents against influenza. However, we think other human cell lines of epithelial, neutrophil, monocyte, and macrophage origins may also be useful for the study of influenza mediated cytokine responses; however, their applications are not as robust for HTS as the U937 cell line and need to be carefully tested and optimized.
CCL2 and CXCL10 were chosen as the readouts for antiinflammatory drug screening against influenza in the U937 cell model for the following reasons: (1) these two factors are closely related to the immunopathology caused by influenza. In the fatal cases caused by an influenza virus infection, the levels of these two cytokines were significantly higher in patients than in healthy controls (de Jong et al. 2006; Deng et al. 2008; To et al. 2010; Gao et al. 2013). (2) Functional studies have shown that knocking out the CCL2 or CXCL10 genes or using neutralizing antibodies to deplete their protein levels and subsequently reducing their functional activity, can significantly alleviate the inflammatory response to influenza infection and improve the survival rate in experimental animal models of influenza infection (Damjanovic et al. 2011; Ichikawa et al. 2013; Wang et al. 2013; Hrincius et al. 2015). (3) CCL2 and CXCL10 are two of the major cytokines/chemokines that recruit monocytes/macrophages and neutrophils, both of which play important roles in influenza-induced inflammation (Ichikawa et al. 2013; Tavares et al. 2017). The reduction of the levels of these two cytokines may decrease the infiltration of inflammatory cells. (4) CCL2 and CXCL10 are regulated by different signaling pathways. CCL2 is primarily regulated by the NFκB pathway whereas CXCL10 is predominantly regulated by the interferon pathway, both of which are key pathways for controlling inflammatory cytokine responses (Chen et al. 2018). (5) Although the inclusion of more cytokines and chemokines may be a better representation of the influenzainduced cytokine storm, too many readouts may unnecessarily complicate the screen. Based on these reasons and concerns, we decided to start with two of the most important pro-inflammatory cytokines, CCL2 and CXCL10.
The use of the U937 cell model to study the anti-inflammatory effects on influenza-induced cytokine responses was validated by testing a panel of known antiviral and immunomodulatory agents, in addition to screening a drug library with 1280 compounds comprised mostly of FDA-approved drugs. Almost all of the immunomodulatory and antiviral agents with known activities in vivo (Table 2) showed expected anti-cytokine and antiviral effects. The screen of the FDA- drug library led to the identification of 16 hit compounds that were shown to inhibit the production of both cytokines, and many hits were shown to inhibit either CCL2 or CXCL10. The average Z' factor for NA, CCL2, and CXCL10 were 0.74, 0.75 and 0.84, respectively, suggesting that the U937 cell model is robust and desirable for HTS of immunomodulatory agents against influenza infection. Interestingly, most of the traditional anti-inflammatory drugs, i.e. steroidal and non-steroid anti-inflammatory drugs, are identified as CCL2 inhibitors. Many novel immunomodulatory agents with new targets were identified in both the anti-CCL2 and anti-CXCL10 hits, covering diverse host targets, including histamine receptors, 5-HT2 receptors, β-adrenergic and cholinergic receptors, HMGCoA reductase inhibitors, DNA gyrase/topoisomerase, ion channels, and tyrosine kinases. It appears that the target distribution of CCL2 and CXCL10 inhibitors are different but related to some extent, suggesting that the expression of CCL2 and CXCL10 are co-regulated by certain signaling pathways, and specifically regulated by their own signaling pathways.
Using the U937 cell model, a secondary screen of the 16 hit compounds identified as inhibiting the expression of both CCL2 and CXCL10 in the primary screen led to the discovery of 6 compounds with IC50 s lower than 10 μmol/L and SIs greater than 20 for anti-cytokine activity. It is worth noting that the 6 re-confirmed hit compounds can be categorized into four classes: (A) TPK inhibitors, (B) Quinolone antibiotics, (C) Statins, and (D) Serotonin reuptake inhibitors. Interestingly, inhibitors targeting these four classes of targets have been previously reported in the literature to reduce inflammation and improve lung pathology caused by influenza infection. The TPK inhibitors, ibrutinib, and imatinib have been reported to protect mice from lethal influenza infection and improve acute lung injury caused by influenza (Hrincius et al. 2015; Florence et al. 2018). The quinolone antibiotic levofloxacin, which has antioxidant effects, increased the survival rate in mice infected with the H1N1 PR8 virus and was shown to inhibit excessive superoxide production in the lungs of mice (Enoki et al. 2015). Statins block the inflammatory signaling pathway and inhibit the production of pro-inflammatory factors in cells. Their use in animal experiments has shown to reduce the mortality of H1N1 and H3N2-infected mice. More importantly, in the retrospective study of human drug use, the use of statins is associated with a reduction in influenza-induced lethality (Liu et al. 2009; Mehrbod et al. 2014; Davidson 2018). Serotonin reuptake inhibitors can alleviate lung inflammation in mice induced by the H1N1 influenza virus and reduce mortality in mice (Sharma et al. 2013). These results indicate that using CCL2 and CXCL10 as readouts in the U937 cell-based model for anti-inflammatory drug screening have predictive value for in vivo efficacy. Of note, many hits that inhibit only CCL2 or CXCL10, with new targets covering HMG-CoA reductase inhibitors, DNA gyrase/topoisomerase IV inhibitors, ion channel inhibitors, and tyrosine kinase inhibitors, may be potentially effective immunomodulators. However, their efficacies need to be further validated.
Conventional cell-based anti-influenza drug screening models typically focus on only one activity, such as antiinflammatory or antiviral activity. Screening anti-inflammatory activity without screening for antiviral effects can be misleading, since antiviral activity itself may lead to a reduction in inflammation (i.e., anti-inflammatory activity). In this study, we have established the U937 cell model to detect both anti-inflammatory and antiviral effects in one assay. This model can be very useful in the identification of the following three types of agents: (1) Agents that only have antiviral effects (2) Agents that only have anti-inflammatory effects. (3) Agents that have both antiinflammatory and antiviral effects.
In summary, we have established a human monocytic cell-based model for high-throughput anti-influenza drug discovery. We have demonstrated that the U937 cell model supports the replication of influenza viruses as well as the production of pro-inflammatory cytokines. Using only three readouts, cytokine CCL2, and CXCL10 expression, and virus NA expression, the U937 cell model can be used to screen immunomodulators and antiviral agents for the treatment of influenza. Because it is cheap and easy to use, the U937 cell model is an important addition to the existing cellular model and the expensive and time-consuming animal models.
-
We thank Dr. Ding Gao and Ms. Juan Min (Center for instrumental analysis and metrology, Wuhan Institute of Virology, CAS) and Professor Xuefang An (Center for the animal experiments) for technical support. This work was supported by the Important Hubei Science and Technology Innovation Plan 2015ACA062 (to Xulin Chen) and the Natural Science Foundation of Hubei Province (2018CFB244, to Jungang Chen).
-
GL and XC designed the experiments. GL, SC, AH, LZ, WS, JC, WT, HZ, CL, and CK performed the experiments. WT, CL, and HZ contributed to the reagents. GL and XC wrote and finalized the manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: