-
Dear Editor,
Chikungunya virus (CHIKV) is a mosquito-borne virus belonging to the family Togaviridae, genus Alphavirus, and was first isolated in Tanzania in the 1950s (Silva and Dermody 2017; Weaver and Lecuit 2015). Human infections with CHIKV typically result in a rapid-onset febrile disease, with symptoms that include fever, headache, rash, severe joint and muscle pain, as well as prolonged periods of disability in some patients (Weaver and Lecuit 2015; Silva and Dermody 2017). Unlike other arboviral diseases such as dengue and Zika fever, the majority of CHIKV infections in humans results in clinical symptoms, with ~15% of human infections who are asymptomatic with seroconversion (Schwartz and Albert 2010; Weaver and Lecuit 2015). CHIKV has been reported in more than 110 countries and territories in Asia, Africa, Europe, and the Americas (CDC 2019), and has evolved into three main lineages, including West African, Asian, and East Central South African (ECSA) (Silva and Dermody 2017; Weaver and Lecuit 2015). The ECSA lineage emerged in Kenya during 2004 and spread to islands in the Indian Ocean, resulting in a large epidemic (Silva and Dermody 2017; Weaver and Lecuit 2015). The first report of CHIKV in China was in 1987 in Yunnan Province (Wu et al. 2012). Since then, several sporadic cases of imported CHIKV infections and a local outbreak have been documented in Guangdong Province (Qiaoli et al. 2012; Wu et al. 2012; Sun et al. 2013; Wu et al. 2013). Here, we describe and present the phylogenetic as well as genetic analysis of a novel CHIKV isolate from a patient with fever and chills traveling from Pakistan to Shenzhen, China in April 2017.
A 49-year-old male patient traveling to Shenzhen from Pakistan was admitted to our hospital on April 30, 2017 with fever and chills, but without headache, arthralgia and rash. Dengue virus (DENV), Zika virus (ZIKV), yellow fever virus (YFV), West Nile virus (WNV), Japanese encephalitis virus (JEV), influenza A virus (IAV) and influenza B virus (IBV) were shown to be negative but positive for CHIKV in the serum screened by commercial kits (Mabsky Biotech Co., Ltd., Shenzhen, China), except for ZIKV which an in-house quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay was used (Yang et al. 2017). Serum samples were then used for subsequent virus isolation and live CHIKV was isolated from C6/36 cells. After three rounds of passage, qRT-PCR was performed with CHIKV-specific primers, and the Ct values of passages 1, 2 and 3 were 33.2, 19.6 and 15.3 (data not shown), respectively. Eleven overlapping fragments covering the entire genome of the isolated CHIKV were amplified with specific primers (Supplementary Table S1) and then sequenced by Sanger sequencing. The assembled sequence was submitted to GenBank with accession no. MH349097 and the strain was designated as SZ-SMGC-1. Meanwhile, electron microscopy of the supernatant showed a cluster of typical CHIKV particles with a diameter ranging from 65.5 to 68.9 nm (Fig. 1A). Interestingly, typical cytopathic effects (CPEs) such as shrinkage, rupture and detachment were found in Vero cell after four days post infection (d.p.i.), while no CPEs were observed in C6/ 36 cells (Fig. 1B). This is consistent with a previous study, which showed that CHIKV infection induced apoptosis in Vero cells but not in C6/36 cells (Wikan et al. 2012). The one-step growth curve of the CHIKV strain was further analyzed in Vero and C6/36 cells. SZ-SMGC-1 replicated to the peak within 36 h post infection (h.p.i.) in C6/36 cell, while it reached the peak at 96 h.p.i. in Vero cell (Fig. 1C). Of note, the peak virus titers in both cells were similar.
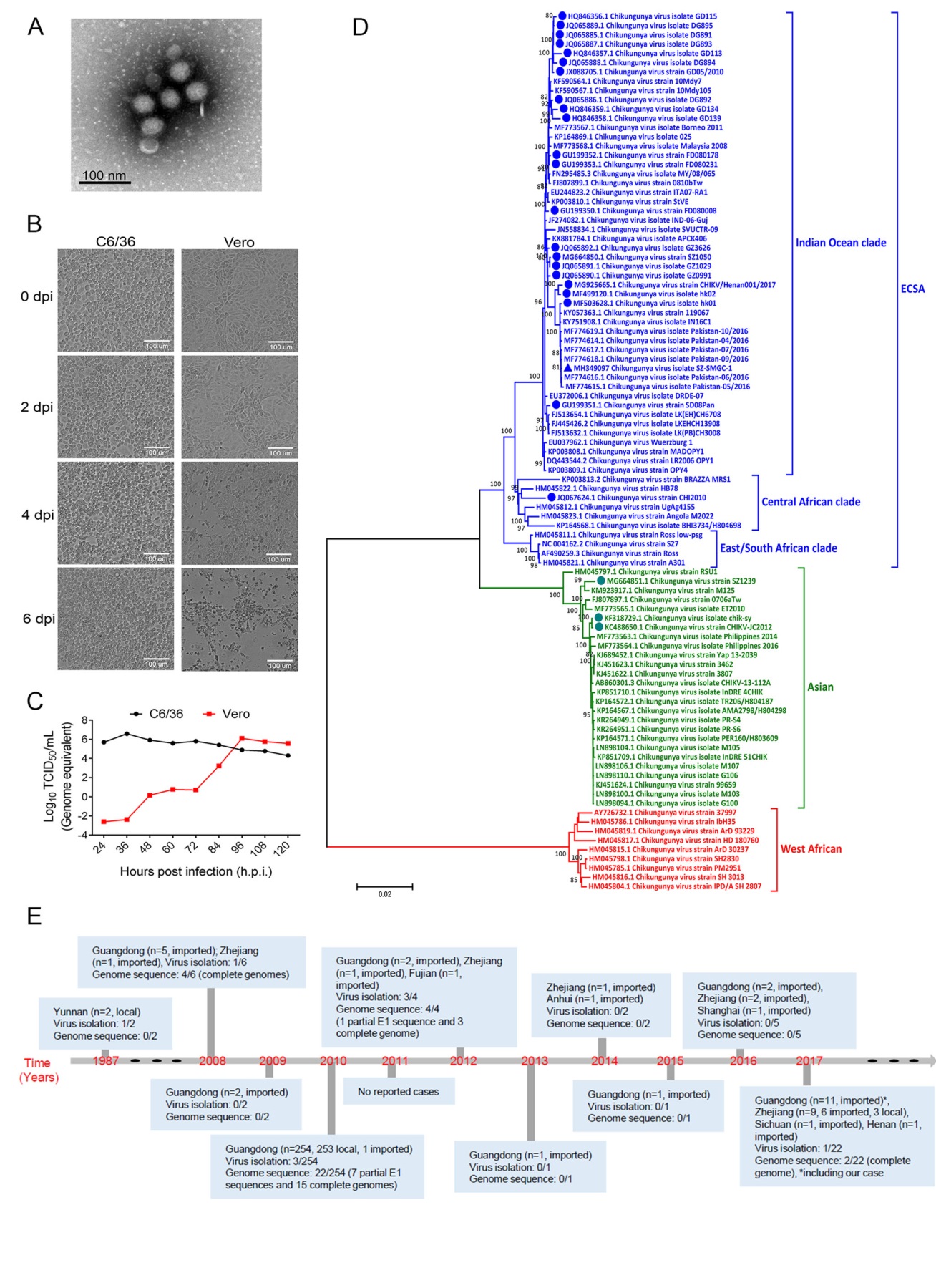
Figure 1. Isolation and characterization of this novel CHIKV strain, and milestones of CHIKV in mainland China, 1987–2017. A C6/36 cells were infected with SZ-SMGC-1, and the supernatant was processed for electron microscopy. A representative image was shown. B C6/36 and Vero cells were infected with SZ-SMGC-1, and the cytopathogenic effects could be observed. C The one-step growth curve of SZSMGC-1 in C6/36 and Vero cells. D Alignment and phylogenetic characteristics of SZ-SMGC-1. SZ-SMGC-1 was indicated with a triangle, and isolates in mainland China were labeled with dots. ECSA: East Central South African. E Milestones of CHIKV in mainland China, 1987–2017.
All CHIKV isolates in China were downloaded and the genetic identities were analyzed (Supplementary Table S2). The nearly complete genome of SZ-SMGC-1 strain showed high nucleotide identity with other isolates in China, ranging from 92.9% to 99.9%, and most closely to the hk01 strain isolated from an imported case into Hong Kong during 2016 (Ho et al. 2017). A total of 95 representative genome sequences of CHIKV were then downloaded from NCBI for phylogenetic analysis (Fig. 1D). Consistent with previous studies (Vignuzzi and Higgs 2017), these CHIKV strains could be classified into three major lineages, including ECSA, Asian and West Africa lineages. Most of the isolates from China (23/26, 88.5%) belonged to the ECSA lineage, including the SZ-SMGC-1; while three (11.5%) isolates belonged to the Asian lineage. The 23 isolates in the ECSA lineage were further clustered into the Indian Ocean and Central African clades. SZ-SMGC-1 closely clustered to viruses identified from Pakistan in 2016, as well as isolates from Henan and Hong Kong in the Indian Ocean clade (Fig. 1D).
Consistent with other CHIKV strains, the genome of the isolated strain contains two ORFs, one encoding the nonstructural protein precursor and the other for the structural protein precursor. Compared with the reference strain S27 (GenBank accession no. NC004162), a total of 38 amino acid substitutions in the non-structural protein and 24 in the structural protein were detected in SZ-SMGC-1, and the substitutions specific for the ECSA lineage (Shi et al. 2017) were shown (Table 1). SZ-SMGC-1 possessed most of the substitutions detected in other strains from China. Furthermore, the D284E and I211T substitutions in E1 and E2 respectively, associated with an increase in transmissibility of CHIKV in A. albopictus (Vignuzzi and Higgs 2017) were also found in SZ-SMGC-1, suggesting that the relative fitness and transmissibility of this SZ-SMGC-1 isolate remains to be characterized in animal or mosquito models. Interestingly, there was an opal stop codon at position 524 of nsp3 protein in SZ-SMGC-1 which was not observed in other closely-related CHIKV strains from Pakistan (Table 1). The nsp3 protein is known to interact with the other three nsps to form the viral replication complex required for viral genome replication, and the majority of CHIKVs possess an opal stop codon at the 3' end of nsp3 protein (Schwartz and Albert 2010; Lum and Ng 2015). A recent study has shown that disruption of the opal stop codon attenuates CHIKV-induced arthritis and pathology, independent of any effects on viral replication (Jones et al. 2017). The presence of CHIKV isolates with the opal termination codon may indicate an evolutionary selection for CHIKV, and the detailed roles of this deletion in CHIKV pathogenesis and replication merit further characterization.
Proteins Substitutionsa Functions SZ-SMGC-1 Pakistan-06 Other strains from China Amino acids No. (%) nsp1 T128K NA K K K 21 (95.5) T 1 (4.5) nsp2 S54N NA N N N 21 (95.5) S 1(4.5) H374Y NA Y Y Y 21 (95.5) H 1 (4.5) L539S NA L L L 12 (54.5) S 10(46.5) nsp3 524 NA Stop codon R Stop codon 20 (909) R 2 (9.1) nsp4 L43A NA A A A 22 (100) L 0 0) R82S NA R R R 12 (54.5) S 10(46.5) T254A NA A A A 21(95.5) T 1(4.5) A 366T NA T T T 21(95.5) A 1(4.5) Q500L NA L L L 22 (100) Q 0 (0) E2 A164T NA T T T 22 (100) A 0(0) I211T Potentiate the ECSA genotype for the finess-increasing effect of A226V T T T 21 (95.5) I 1 (4.5) K252Q Enhance the infection eficiency to A. albopictus K K K 10 (46.5) Q 12 (54.5) V386A NA A A A 21 (95.5) V 1 (4.5) E1 S72N NA N N N 22 (100) S 0(0) E211K NA E E E 7 (31.8) K 15 (68.2) A226V Increased fitness to the A. albopictus A A A 10 (46.5) V 12 (54.5) D284E Influence the icosahedral E1-scaffold and increase transmissibility E E E 21 (95.5) D 1 (4.5) NA not available, ECSA East Central
aSouth African."Specific substitutions for viruses from ECSA lineage were shown.Table 1. Key amino acid substitutions of the SZ-SMGC-1 and the other isolates in China.
Currently, most cases of CHIKV in China were reported in Guangdong Province, followed by Zhejiang Province (Fig. 1E), two areas with frequent external contacts. In 2010, an outbreak of CHIKV in Dongguan, Guangdong Province, resulted in 253 human infections (Qiaoli et al. 2012; Wu et al. 2012). Interestingly, phylogenetic analysis of these sequences from Dongguan showed the highest homology with a Thailand strain (GenBank accession no. FJ882911) isolated in 2009 (Wu et al. 2012), suggesting that this outbreak may have been caused by importation of the asymptomatic or undiagnosed/undetected CHIKV patients. The main vectors for CHIKV are A. aegypti and A. albopictus mosquitoes (Silva and Dermody 2017; Weaver and Lecuit 2015), which are widely distributed in southern and eastern China (Weaver and Lecuit 2015; Xia et al. 2018). Currently, no licensed CHIKV vaccines are available (Weaver and Lecuit 2015; An et al. 2017), thus the wide distribution of CHIKV vectors and human population without pre-existing immunity against CHIKV means that there is a risk of the virus re-emerging in these areas. Therefore, staff in the borders/ports and clinicians should pay more attention to travelers with Chikungunyalike clinical features, especially those from regions known to be endemic for CHIKV.
HTML
-
This work was supported by the grants from the National Natural Science Foundation (NSFC) of China (81802004), the Shenzhen Science and Technology Research and Development Project (JCYJ20160427153238750), the National Science and Technology Major Project (2018ZX10711001, 2017ZX10103011), Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (XDB29010102), the Sanming Project of Medicine in Shenzhen (SZSM20141 2003). Y.B. is supported by the NSFC Outstanding Young Scholars (31822055).
-
All procedures performed in studies involving human participants were in accordance with the guidelines approved by the Ethics Committees from Shenzhen Third People's Hospital (SZTHEC2016001) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from the participant enrolled in the study.
















 DownLoad:
DownLoad: