HTML
-
Antibodies, the essential components of humoral immunity, are a major defense line against viral infections. To neutralize viral infections, antibodies primarily bind to specific epitopes on the outer surfaces of viral particles. An important first step in modern structural vaccinology involves structurally characterizing the interactions occurring between viral antigens and their cognate antibodies (Anasir and Poh 2019). This step provides the direct evidence for the location of viral epitopes, which helps to elucidate the neutralization mechanisms of antibodies. The first high-resolution structure of a virus (tomato bushy stunt virus) was solved four decades ago using X-ray crystallography (Harrison et al. 1978), a technique that has been primarily limited to determining the structures of relatively simple non-enveloped viruses (http://viperdb.scripps.edu/xray.php) or viral components. Because crystallizing virus-antibody complexes can be challenging, morphological studies of virus-antibody interactions have long been carried out through transmission electron microscopy (TEM) with negative staining protocols (Almeida and Waterson 1969). The resolution achieved by this technique is usually low, but the structural details can be enhanced by modeling high-resolution crystal structures into low-resolution TEM maps. As the negatively-stained samples are visualizable with a conventional TEM instrument, they are still widely used today for characterizing immune complexes with human viruses like influenza A (Ekiert et al. 2012), Marburg (Flyak et al. 2015) and Ebola viruses (Flyak et al. 2016).
Cryo-electron microscopy (cryo-EM), a Nobelprize-winning technique, is now routinely used for studying virus-antibody complexes at high resolution (Earl and Subramaniam 2016). In contrast to negatively-stained samples, which might be significantly distorted by the dehydration process and the presence of stains, virus particles in cryo-EM studies are freshly frozen in a thin layer of vitreous ice to maintain their native conformations, making high-resolution analyses possible. However, before direct electron detection cameras (DEDs) were introduced in 2012, cryo-EM structures could only be solved at subnanometer resolutions with traditional CCD cameras, except some pioneer work based on images recorded on photographic film (Zhang et al. 2010). Largely resulting from the development of DED technologies, better microscopes and sophisticated reconstruction algorithms, cryo-EM has developed in recent years as a powerful highresolution technique in structural biology research (Shen 2018).
There are two major 3D cryo-EM analysis strategies: single particle analysis (SPA) and cryo-electron tomography (cryo-ET) (Danev et al. 2019). SPA is usually used for macromolecular assemblies that are stable, soluble and homogeneous in vitro. Because some viruses are highly structurally ordered, especially those with icosahedral symmetries, it is possible to solve the structures of whole viral particles at better than 3 Å resolution using SPA by combining data from several thousands of purified virus particles (Jiang and Tang 2017). The density maps at this resolution allow the de novo building of atomic models. For viruses with pleomorphic shapes, 3D reconstruction of a single vitrified virion is achievable using cryo-ET procedures at ~25-Å resolution. Sub-tomogram averaging techniques are further exploitable for achieving better structural details for repeating structures, such as the surface glycoproteins on viruses. Both SPA and cryo-ET approaches have been used for structural studies on the interactions occurring between whole virions (or viral components) and their associated antibodies.
-
Antibody molecules for cryo-EM studies can be generated in different ways. Mass production of murine monoclonal antibodies (mAbs) has traditionally been achieved by the use of hybridoma technology where antibody-producing B cells derived from immunized mice are fused to an immortalized cell line (e.g., myeloma) (Kohler and Milstein 1975). Recombinant antibodies can be generated by phage display technology followed by in vitro selection, which targets the whole virus particles or some viral components (Hoogenboom 2005). Various methods for isolating antigen-specific human B cells to obtain mAbs have also been reported (Crowe 2017). For viruses with naturally occurring high mutation rates like HIV-1, broadly neutralizing antibodies (bNAbs) that potently target a wide range of viral strains have been isolated from virus-infected individuals and extensively studied using cryo-EM (Stephenson and Barouch 2016; Chuang et al. 2019).
For cryo-EM analyses, the antibody components are IgG molecules or their truncated parts. In a single IgG molecule, two antigen binding fragments (Fabs) are present (Fig. 1A, 1B), a situation that could result in highly heterogeneous antigen–antibody complexes if one antigen particle becomes crosslinked with another. Except for SPA studies on IgG bivalency (Ye et al. 2016), IgG is usually used in cryo-ET studies at medium resolution for HIV-1 (Tran et al. 2012), influenza (Tran et al. 2016b) and Ebola viruses (Tran et al. 2016a). Compared with the intact IgG molecule, the monovalent Fab fragments generated by papain digestion of whole IgG molecules are more commonly used for structural studies on virus-antibody complexes (Tables 1, 2, 3). The single-chain variable fragment (scFv) generated by fusing the variable regions of the heavy (VH) to the light (VL) chains with a flexible peptide linker (Finlay et al. 2017) can be used for cryo-EM studies of virus-antibody interactions (Kaufmann et al. 2009; Liu et al. 2017). Single-domain antibodies (sdAbs), such as variable domains of heavy chain-only antibodies (termed VHH) and VH domains of human IgG molecules, have also served as antibody derivatives for cryoET studies when combined with sub-tomogram averaging (Meyerson et al. 2013).
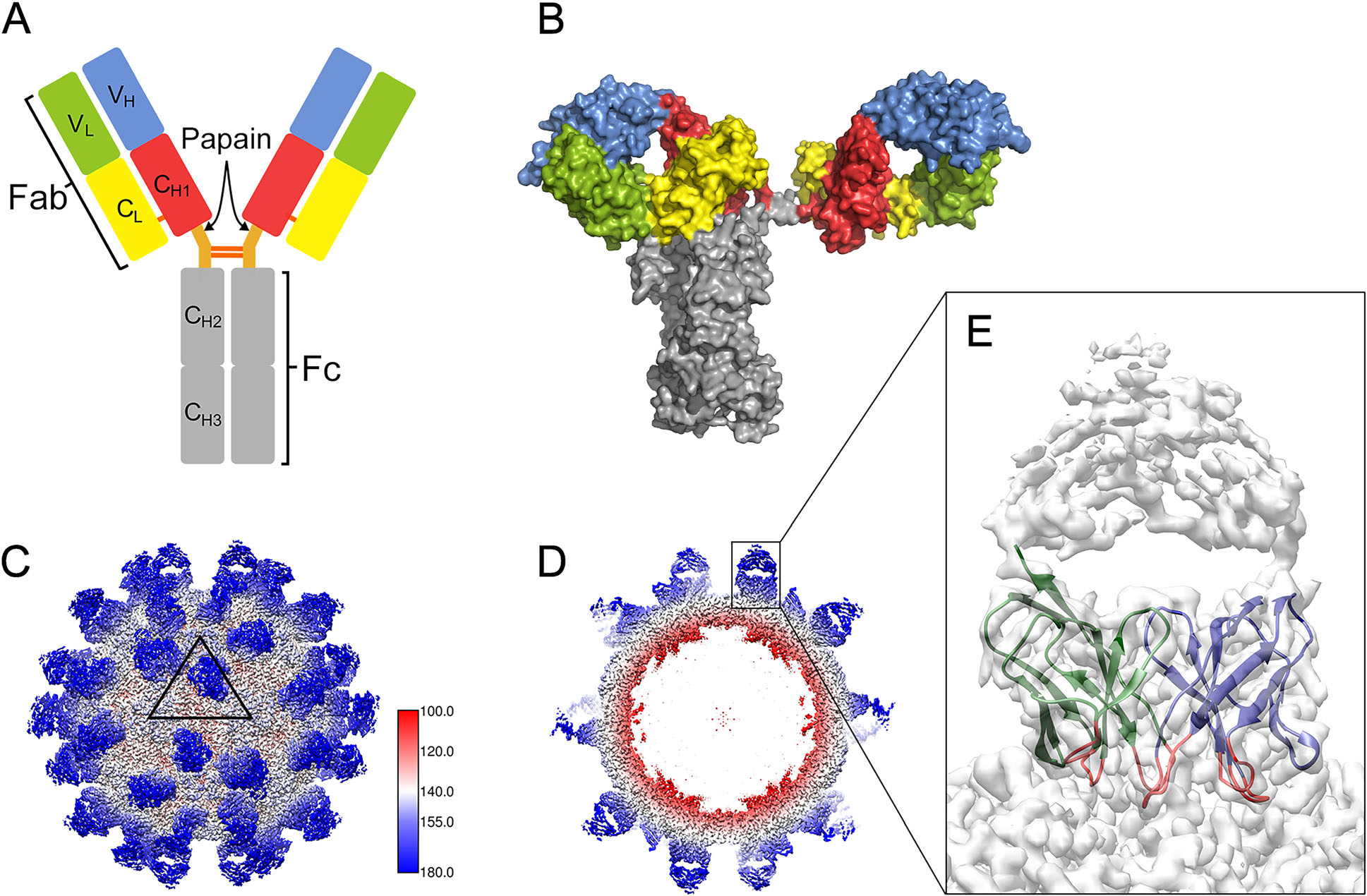
Figure 1. Structural models of antibody molecules on the viral surface. A Modular organization of a Y-shaped IgG molecule, which consists of two heavy chains and two light chains covalently linked via disulfide bonds. Papain digestion of a parental antibody produces two Fab fragments and one Fc fragment. A Fab fragment comprises VH, CH1, VL and CL domains. B Surface illustration of highly asymmetric human IgG1 b12 (PDB-1HZH) colored by domain as in (A). C CryoEM map of Fab AT12-015 complexed with HPeV3 (EMD-0069), showing that the 60 copies of Fab (blue) are placed on the viral surface (white). A triangular icosahedral asymmetric unit (ASU) is outlined on the capsid surface. The numbers show the positions of neighboring fivefold and two threefold axes limiting the ASU. D Central cross-section of the cryo-EM map showing the Fab densities (blue). E The enlarged inset shows the density corresponding to one Fab AT12-015 molecule. The VH and VL domains represented by green and blue ribbons are well defined in the density map. The six complementarity-determining regions in VH and VL are colored red.
Genus Virus name Antigen Antibodya Fragment EMD codes Resolution
(Å)Enterovirus Coxsackievirus (CV) CV-A6 A-particle 1D5 Fab 6757 3.8 CV-A10 A-particle 2G8 Fab 9603 4.3 CV-A10 mature virion 2G8 Fab 9604 3.9 CV-A10 procapsid 2G8 Fab 9605 4.2 Enterovirus (EV) EV-A71 mature virion MA28-7 Fab 5673 23.4 E18 Fab 2397 10 E19 Fab 2436 13 D5 IgG 6365 7.2 D5 Fab 6366 4.8 EV-A71 procapsid D5 Fab 6383 6 22A12 Fab 6200 8.8 E18 Fab 2434 16 D6 Fab 6963 4.9 A9 Fab 6964 6.8 EV-A71 VLP D5 IgG 6384 5.5 EV-D68 mature virion 15C5 Fab 9633 3.6 15C5/11G1 Fab 9634 3.5 EV-D68 A-particle 11G1 Fab 9636 7.2 Poliovirus (PV) PV1 mature virion A12 Fab 5670 12 C3 Fab 5291 11.1 PVSP6A VHH 5886 4.8 PVSP29F VHH 5888 6.5 PVSS8A VHH 6433 4.2 PVSP19B VHH 6434 4.8 PVSS21E VHH 6435 3.8 PV2 mature virion A12 Fab 5671 20 PV1 procapsid P1 Fab 5283/5284/5285/5286 13/21/18/18 C3 Fab 5293 22 PV1 A-particle P1 Fab 5280/5282 12/26 C3 Fab 5292 9.1 PVSP17B VHH 8285 5.3 PVSS12B VHH 8285 5.3 PVSS10E VHH 8277 4.8 PVSS7A VHH 8286 5.3 Rhinovirus (RV) RV B14 mature virion C5 Fab 8754/8761/8762 2.53/2.71/2.26 RV B14 procapsid C5 Fab 8763 3.01 Aphthovirus Foot-and-mouth disease virus (FMDV) FMDV-O mature virion D9 Fab 0173 3.97 Parechovirus Human parechovirus (HPeV) HPeV-3 mature virion AT12-015 Fab 0069/3138 2.8/15 HPeV-1 mature virion AM28 Fab 2761 19.76 Hepatovirus Hepatovirus A (HAV) HAV mature virion R10 Fab 6688 4.2 F4 Fab 9827 3.9 F6 Fab 9828 3.68 F7 Fab 9829 3.05 F9 Fab 9830 3.79 aStructural insights into the possible mechanisms for antibody-mediated neutralization discussed in the text are summarized below. 1D5: inhibition of virus-cellular binding ( Xu et al. 2017 ), 2G8: capsid stabilization (Zhu R et al. 2018 ), MA28-7: cross-linking of virions and blocking receptor binding (Lee et al. 2013 ), E18: induction of genome release (Plevka et al. 2014 ), D5: capsid stabilization (Ye et al. 2016 ), 22A12: capsid stabilization (Shingler et al. 2015 ), D6: blocking receptor binding (Zhu L et al. 2018b ), A9: blocking receptor binding and capsid destabilization (Zhu L et al. 2018b ), 15C5: blocking receptor binding and locking capsid at intermediate stage, 11G1: locking capsid at intermediate stage (Zheng et al. 2019 ), R10: blocking receptor binding (Wang X et al. 2017 ), F4, F6, F7 and F9: blocking receptor binding (Cao et al. 2019 ).Table 1. Summary of cryo-EM structures from picornavirus-antibody complexes in the Electron Microscopy Data Bank (EMDB) (https://www.ebi.ac.uk/pdbe/emdb/).
Virus name Antigen Antibodya Fragment EMD codes Resolution (Å) Dengue virus (DENV) DENV1 mature virion 14c10 Fab 5268 7 1F4 Fab 2442 6 DENV2 mature virion 747(4)B7 Fab 2818 10.24 1A1D-2 Fab 1418 24 2D22 Fab 2967/2968/2969/2996/2997/2998/2999 6.5/20/21/6.9/13/11/23 DENV2 immature virion 2H2 Fab 5674/5675/5676/5677 21/25/21/21 E53 Fab 5102 23 DENV3 immature virion 1H10 Fab 9649/9650/9651 12/25/25 DENV3 mature virion 5J7 Fab 5935 9 West Nile Virus (WNV) immature virion E53 Fab 5103 15 Mature virion E16 Fab 1234 14.5 E16 scFv 5115 22.75 CR4354 Fab 5190 13.7 Tick-borne encephalitis virus (TBEV) Mature virion 19/1786 Fab 3754/3755 3.9/19.2 Zika virus (ZIKV) Mature virion ZIKV-117 Fab 8548 6.2 ZKA190 Fab 6793/6794 22/22 Z23 Fab 9542 9.4 C10 Fab 9573/9574/9575 4.4/12/4 ZAb-FLEP Fab 7613 9.7 ZK2B10 Fab 9811/9812 20/11 ZIKV-195 Fab 9131 4 Japanese encephalitis virus (JEV) Mature virion 2F2 Fab 6854 4.7 2H4 Fab 6855 4.6 aThe possible neutralization mechanisms for flavivirus antibodies discussed in the text are summarized below. 14c10: blocking receptor binding ( Teoh et al. 2012 ), 1F4: blocking virus attachment (Fibriansah et al. 2014 ), 1A1D-2: blocking virus attachment by binding to hidden epitopes (Lok et al. 2008 ), 2D22: blocking capsid reorganization required for virus fusion (Fibriansah et al. 2015a ), 2H2: inhibition of virus maturation (Wang et al. 2013 ), E53: binding to partially immature heterogeneous virions (Cherrier et al. 2009 ), 1H10: enhancing immature virus attachment to endosomal membrane (Wirawan et al. 2019 ), 5J7: blocking receptor binding and capsid stabilization (Fibriansah et al. 2015b ), E16: blocking capsid reorganization required for virus fusion (Kaufmann et al. 2006 ), ZIKV-117: capsid stabilization (Hasan et al. 2017 ), ZKA190: inhibition of either cell attachment or membrane fusion (Wang J et al. 2017 ), C10: capsid stabilization (Zhang et al. 2016 ).Table 2. Summary of cryo-EM structures from flavivirus-antibody complexes.
Genus Virus name Antigen Antibody Fragment EMD codes Resolution (Å) Lentivirus Human immunodeficiency virus (HIV) HIV-1 BaL virion A12 VHH (Tomo) 5544/5551 m36 VH (Tomo) 5552/5553/5554/5555 17b IgG (Tomo) 5456 22 VRC01 IgG (Tomo) 5457 24 VRC03 IgG (Tomo) 5458 23 VRC02 Fab (Tomo) 5459 23 VRC02 IgG (Tomo) 5460 25 VRC01/17b IgG (Tomo) 5461 28 b12 Fab (Tomo) 5018/5021 20/20 17b Fab (Tomo) 5020/5023 20/20 17b/ A32 Fab 0466 13.08 BG505 SOSIP.664 17b/8ANC195 Fab 7516 /(Tomo) 3096 3.54/23 3BNC117 Fab 8644 4.4 3BNC117/ PGT145 Fab 8643 4.3 3BC315 Fab 3067 9.3 BG1/8ANC195 Fab 8693 6.2 PG9/8ANC195 Fab 8695 11.5 3417 Fab 7552/7553/7554/7555/7556/7557 4.7/4.7/4.7/4.7/4.7/4.7 VRC34.01 Fab 8125 17 BF520.1 Fab 9166 4.8 PGT128 Fab 3121/3120 4.36/4.47 17b Fab 8730 8.6 PGV04 Fab 5779/5780/5781 5.8/7.9/8.2 PGT151 Fab 9062 4.5 BG505 DS- SOSIP.664 vFP/VRC03/PGT122 Fab 7622/7621/7459/7460 4/4/3.8/3.6 vFP Fab 8420/8421/8422 8.58/14.7/19.6 PGT145 Fab 8427 6.8 2G12/ VRC03 Fab 8981 8.8 PGT122/VRC03/ FP antibodies Fab 9189/20189/20191/9359/9320/9319/8977 3.8/4.3/3.5/3.7/4.2/4/3.18 462c SOSIP.664 VRC01GL Fab 9294/9295/9303 /9304 3.8/3.8/4.8/4.8 B41 SOSIP. 664 17b Fab 8713 3.7 PGV04 Fab 8716 7.4 b12 Fab 8717 3.6 21c/8ANC195 Fab 9038 4.06 PGT151 Fab 9030 6.7 ZM197 SOSIP. 664 VRC01 Fab 3059 9.32 PC64M18C043 FL Env PGT151 Fab 7858 3.1 PGT151/PCT64-35S Fab 7859 6.8 PC64M18C043 SOSIP. 664 PGT151 Fab 7860 4.9 PC64M4C054 SOSIP. 664 PCT64-13C Fab 7863/7864/7089 5.1/30/13.2 PCT64-13F Fab 7862 30 PCT64-35S Fab 7865/7866 5.5/8.2 PC64M4C054 FL Env PGT151/PCT64-13C Fab 7861 30 JR-FL EnvΔCT PGT151 Fab 3308/3309 4.19/4.3 PGT151/10E8 Fab 3312 8.8 AMC011 SOSIP.v4.2 PGV04 Fab 8302 6.2 KNH1144 SOSIP. gp140 VRC03 Fab 2484 6 17b Fab (Tomo) 5462 8.8 Lymphocryptovirus Epstein-Barr virus (EBV) glycoprotein AMMO1 Fab 7344/7345 4.8/10 Betacoronavirus Middle East respiratory syndrome-related coronavirus (MERS-CoV) S protein G4 Fab 8783/8784/8785/8786/8787/8788/8789/8790/8791/8792/8793 4/3.6/4.8/4.6/4.8/4.7/5/4.5/4/4/11.5 LCA60 Fab 0401/0402 3.5/3.6 Severe acute respiratory syndrome coronavirus (SARS-CoV) S protein S230 Fab 0403/0404 4.2/4.5 Alphainfluenzavirus Influenza virus Influenza virion 6F12 IgG (Tomo) 6610/6611 25/25 C179 IgG (Tomo) 5684/5685 7B2 IgG (Tomo) 6612 25 3F5 IgG (Tomo) 6613/6614 25/25 HA protein K1915 scFv 8561/8562/8563/8564 4.8/4.8/4.8/4.8 H7.5 Fab 9142/9143/9145 7.4/9.2/7.4 Ebolavirus Ebola virus (EBOV) glycoprotein 100/114 Fab 3310/3311 7.2/6.7 c2G4/c13C6 IgG/Fab 8240 4.3 c13C6/BDBV91 IgG/Fab 8241 5.5 c4G7/c13C6 IgG/Fab 8242 4.3 ADI-15878 Fab 8935/8936 4.14/4.29 VLPs c13C6 IgG (Tomo) 8226 25 c2G4 IgG (Tomo) 8227 25 c4G7 IgG (Tomo) 8228 25 Table 3. Summary of cryo-EM structures from glycoprotein–antibody complexes.
Thus far, 3D reconstructions of virus-Fab complexes at atomic resolution are available for some picornaviruses, including rhinovirus B14 (RV-B14) (Dong et al. 2017), human parechovirus 3 (HPeV3) (Domanska et al. 2019), hepatovirus A (HAV) (Cao et al. 2019) and enterovirus D68 (EV-D68) (Zheng et al. 2019) (Table 1). The atomic models of the capsid protein were well-fitted in the cryoEM density map for HPeV3 (Fig. 1C, 1D). Nevertheless, only the VH and VL domains have clearly defined densities (Figs. 1E, 2G), whereas the density regions in the CH1 and CL domains are less ordered, reflecting the flexibility of the linker regions between the constant domains (CH1 and CL) and variable domains (VH and VL). Because the antigen binding site is determined by six hypervariable loops, namely, the complementarity-determining regions (CDRs) on the VH and VL domains, these densities can provide detailed information about the virus-antibody interface. Based on high-resolution structural analysis, CDRs from five HAV antibodies (R10, F4, F6, F7 and F9) interact with a single conserved antigenic site, which has been shown as an attractive target for rational development of antiviral drugs (Wang X et al. 2017; Cao et al. 2019).
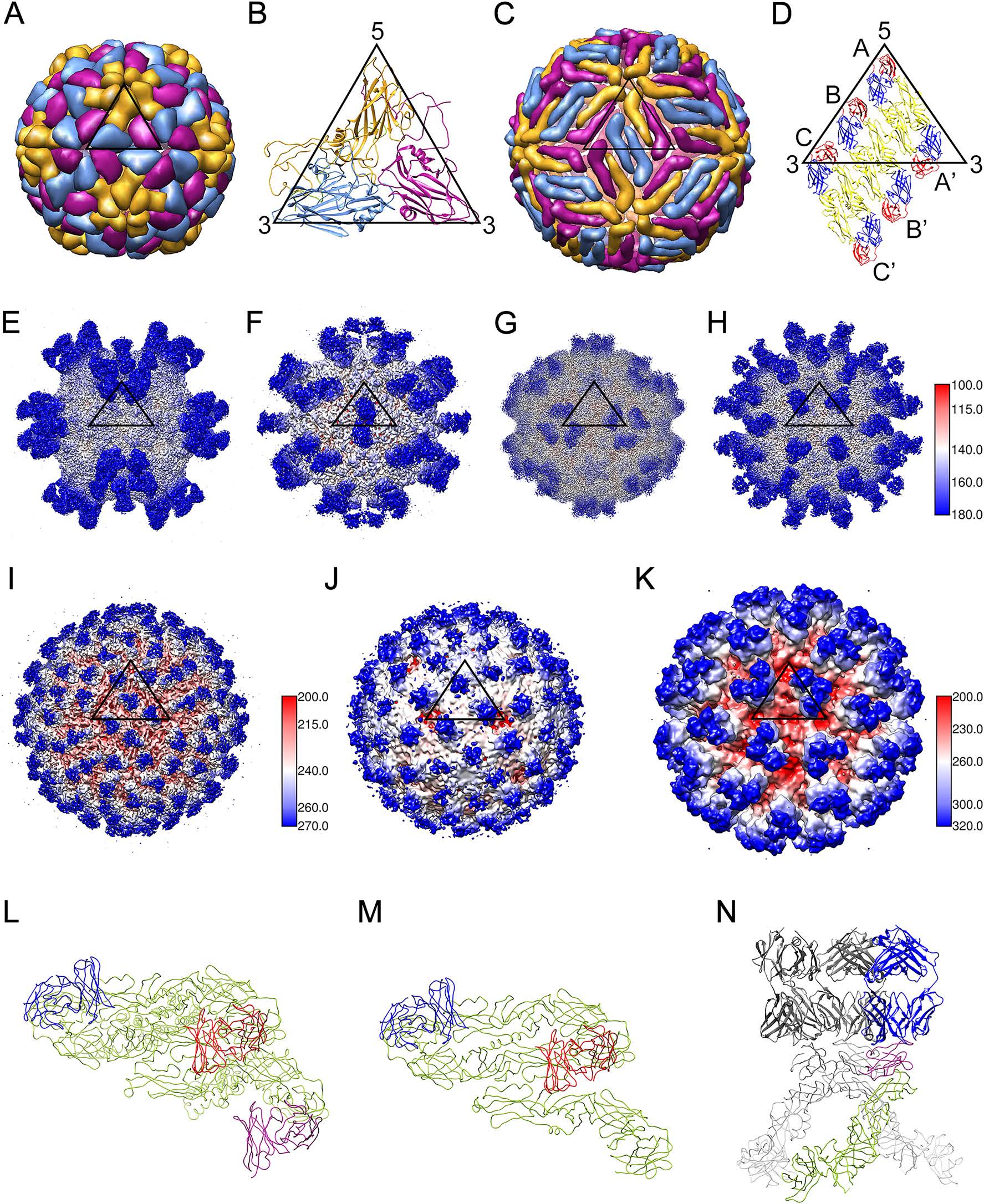
Figure 2. Virus-antibody complexes from picornaviruses (T = pseudo 3) and flaviviruses (T = 3). A The capsid shell on human rhinovirus 14 (PDB-4RHV) is formed by 60 copies of VP1 (gold), VP2 (blue), and VP3 (magenta). B In an ASU, VP1, VP2 and VP3 are folded with a similar "jelly-roll" topology. C The dengue virus capsid structure (PDB-1K4R) is shown as a smooth herringbone lattice of 90 dimers ('E dimers'). The three E molecules in an ASU are colored gold, blue and magenta. D E dimers in two ASUs. The E protein's ectodomain has three domains: DI (red), DII (yellow) and DIII (blue). E–H CryoEM structures of Fab–picornaviruses complexes, where 60 copies of Fab (blue) bind to the outermost surface of the virus (white) near the 5-, 2-, 3- and q3-fold vertices (5-fold: EMD-6757, 2-fold: EMD-6366, 3-fold: EMD-8762 and q3-fold: EMD-9604, respectively. See Table 1 for related information). An icosahedral ASU is outlined in each map. I, L 180 copies of Fab 2D22 (blue) bind to DENV2 at 4 ℃ (EMD- 2967). J, M 120 copies of Fab 2D22 (blue) bind to DENV2 at 37 ℃ (EMD-2968). K, N 180 copies of Fab 2H2 bind to immature DENV2 (EMD-5674). There are 60 characteristic spikes on the immature virion. Each spike is a hetero-hexamer consisting of three prM (magenta) and three E molecules (green). Virions are radially colored as in the side bar with the number corresponding to the radius (in Å). Note that the picornavirus and flavivirus particles are not drawn to scale. The external diameters of picornaviruses and flaviviruses are ~ 30 nm and ~ 50 nm, respectively.
-
Viruses with icosahedral capsid shells are the most studied antigens in immune complexes by cryo-EM techniques, largely due to their high symmetry. Since perfect symmetry could be broken as observed in many situations, such as symmetry mismatches at the portal vertex of herpesviruses (Parent et al. 2018) and partially mature particles of dengue viruses (Rodenhuis-Zybert et al. 2011), block-based or localized reconstruction strategies are now often applied to icosahedral particles showing conformational flexibility or symmetry mismatches (Ilca et al. 2015; Zhu D et al. 2018). Geometrically, an icosahedral object has 60 equivalent positions related by fivefold, threefold and twofold rotational symmetry. The 60 repeating units occupying each of these positions are referred to as asymmetric units (ASUs). The triangulation number (or T-number) is usually assigned to an icosahedral virus to identify the number of subunits in an ASU. In the simplest T = 1 virus (e.g., parvoviruses), only one capsid molecule is present in the ASU, and the 60 capsid protein copies form an enclosed shell to protect the viral genome. In contrast, in a T = 3 virus (e.g., flaviviruses), the ASU consists of three chemically identical capsid proteins (e.g., the E protein from flaviviruses), each of which undergoes conformational adjustments to occupy three "quasi-equivalent" positions in the ASU. Some icosahedral viruses show "pseudo" T geometry. Picornaviruses (T = pseudo 3) have an ASU of three capsid protein types (VP1, VP2 and VP3), possibly making them a special T = 1 case. Next, we will examine the epitope distribution on these two types of humaninfection viruses, and also discuss the binding capacity of Fab molecules on their virions.
Small non-enveloped viruses in the Picornaviridae family cause mild or severe human infections, and provide important antigenic particles for cryo-EM studies (Table 1). Crystal structures of rhinovirus (Rossmann et al. 1985) and poliovirus (Hogle et al. 1985) have revealed that on the icosahedral capsid shell, VP1 lies close to fivefold axes and the neighboring VP2 and VP3 are found around threefold axes (Fig. 2A, 2B). Since 1997, outbreaks of hand-foot-and-mouth disease have been increasingly reported, the leading pathogens of which are various enteroviruses, including enterovirus A71 (EV-A71), coxsackievirus A6 (CV-A6), CV-A10 and CV-A16 (Kimmis et al. 2018; Yu and Cowling 2019). Crystallographic and cryo-EM studies have shed some light on the uncoating mechanism of EV-A71, CV-A10 and CV-A16 (Wang et al. 2012; Ren et al. 2013; Zhu L et al. 2018a). Based on the cryo-EM structures of the enterovirus-Fab complexes, four neutralizing sites have been identified (Fig. 2E–2H) (Zhu R et al. 2018). Site 1 is located near the icosahedral fivefold axis of EV-A71 (MA28-7) (Lee et al. 2013), CV-A6 (1D5) (Xu et al. 2017) and EV-D68 (11G1) (Zheng et al. 2019). While site 2 maps to the VP1 GH-loop across the twofold axis of EV-A71 (22A12 and D5) (Shingler et al. 2015; Ye et al. 2016), site 3 is situated near the threefold axis of EV-A71 (E18, E19 and A9) (Plevka et al. 2014; Zhu L et al. 2018b) and EV-D68 (15C5) (Zheng et al. 2019). Site 4 is adjacent to the quasi threefold axis of CV-A10 (2G8) (Zhu R et al. 2018).
Some clinically relevant viruses in the Flaviviridae family, such as dengue virus (DENV), West Nile virus (WNV), Japanese encephalitis virus (JEV), tick-borne encephalitis virus (TBEV) and Zika virus (ZIKV) (Holbrook 2017; Yang et al. 2019), have been structurally studied as antigenic particles using cryo-EM (Table 2). In contrast with non-enveloped enteroviruses, flaviviruses possess host-derived lipid bilayers with 180 pairs of envelope (E) and membrane (M) proteins on the viral membrane (Perera and Kuhn 2008). Organized as head-totail homodimers on the outer surface (Fig. 2C, 2D), the E protein plays important roles in receptor binding and in mediating virus-host membrane fusion. Compared with picornaviruses, flavivirus epitopes do not possess an apparent pattern near symmetrical axes (Fig. 2I, 2J). Based on how Fab interacts with the E dimer, different epitopes on the DENV E protein are classifiable into four groups: (1) those that occur within an E monomer (e.g., 1F4) (Fibriansah et al. 2014); (2) those spanning the adjacent surface of two E molecules from neighboring E dimers (e.g., 14c10) (Teoh et al. 2012); (3) those consisting of amino acid residues from the two E molecules within an E dimer (e.g., 747(4)B7) (Dejnirattisai et al. 2015); and (4) those that occur across three neighboring E molecules (e.g., 5J7) (Fibriansah et al. 2015b). Largely based on antibody-E complex crystal structures, antibodies may target DI domain, DII fusion loop epitope (FLE) or DIII domain within an E monomer (Dai et al. 2016).
The distance between neighboring epitopes can impact the number of bound antibody molecules on each virion. Although both MA28-7 and 1D5 Fabs bind to site 1 of picornaviruses, only one MA28-7 Fab fragment occupies each fivefold vertex (Lee et al. 2013) while five 1D5 Fab molecules bind each fivefold vertex of CV-A6 (Xu et al. 2017). Compared with 1D5, Fab MA28-7 is closer to the symmetry axis, which renders steric hindrance between possible Fabs, thereby limiting the number of bound Fabs. As another example, the bivalent binding pattern of D5 was characterized in which the two Fab IgG fragments could bind to the GH loops of neighboring VP1 molecules related by twofold symmetry, a finding consistent with the observation that D5 IgG was able to neutralize EVA71 much more potently than D5 Fab (Ye et al. 2016). Contrastingly, the 22A12 binding sites near twofold axes on EVA71 are further apart and bivalent binding of an antibody cannot occur (Shingler et al. 2015). In some cases, Fab binding can even change the local arrangement of the E protein to accommodate more Fab molecules. For example, when the total 180 copies of Fab ZKA190 bind to the ZIKV surface, E proteins at the fivefold vertices move apart and steric clash is avoided (Wang J et al. 2017).
Structural variations in the capsid protein at quasiequivalent positions may also impact the number of bound Fab molecules. Within an ASU, the DENV E protein exists as three conformations showing slight structural variation. 180 copies of Fab 747(4)B7 in total can bind to a DENV virion (Dejnirattisai et al. 2015), suggesting that such variations have no apparent impact on the epitope. However, in other cases, conformational changes can result in a less effective epitope. For example, Fab 1F4 (targeting the DI and DI–DII hinges) does not bind to the E proteins near the threefold vertices where the epitope is partially hidden; consequently, only 120 copies of Fab 1F4 interact with a DENV virion (Fibriansah et al. 2014). Additionally, the capacity of Fab to bind onto a virus particle may not be straightforward when the binding sites are not fully occupied, as shown in the density analysis of ZIKV-117 Fab in a cryo-EM reconstruction (Hasan et al. 2017).
-
Antibodies have been used to capture intermediate states in the assembly pathway of enteroviruses. There are two major enterovirus particles in infected host cells: empty procapsids (noninfectious) and mature virions (infectious). Upon binding to cellular receptors, the native virions are converted into uncoated intermediates called A(altered)- particles (Shingler et al. 2013). Binding to the E18 antibody transforms infectious EV-A71 into A-particles and triggers genome release (Plevka et al. 2014). Uniquely, CV-A6 A-particles are biochemically and structurally stable, which enabled the A-particle-Fab complex to be reconstructed at a 3.8-Å resolution (Xu et al. 2017). 2G8 shows cross-reactivity against the CV-A10 procapsid, the mature virion, and the A-particle, suggesting that the epitopes on 2G8 are structurally conserved among the three capsid forms (Zhu R et al. 2018).
The dynamic conformational changes occurring during the flavivirus life cycle have also been investigated by cryoEM. Differing from the mature virus particles with smooth surfaces, immature virions appear as rough particles with 60 spikes comprising three E and three prM molecules (the pr peptide on top of each trimeric spike) (Perera and Kuhn 2008). As the pr peptide is cleaved during virus maturation and is absent in mature virions, anti-prM Fabs (e.g. 2H2 and 1H10) form complexes with immature DENV (Fig. 2K, 2N) (Wang et al. 2013; Wirawan et al. 2019). Highly crossreactive E53, a fusion-loop-specific antibody, binds preferentially to spikes on immature DENV and WNV particles (Cherrier et al. 2009). Because these antibodies can trap flaviviruses in immature states, the neutralizing mechanism for them may depend on their capacity to block the normal transition occurring during the maturation process.
Many cryo-EM studies have been performed to investigate antibody-virus particle interactions for different functional states, including the following ones: (1) Intermediate complexes during 'breathing' motion. Although each cryo-EM reconstruction usually represents a static snapshot of a specific conformation, structural plasticity in immune complexes can also be visualized. Fab 1A1D-2 induces large conformation changes in the E protein and binds to normally partially hidden epitopes (Lok et al. 2008). Cryo-EM studies have also revealed that Fab 1A1D-2 only binds to epitopes near the fivefold and threefold vertices, suggesting that the extent of the breathing might be not evenly distributed over the viral surface. (2) Size variations in the viral shells at different temperatures. It was found that when exposed to 37 ℃, DENV virions expand in size when compared with the structure at 4 ℃ (Fibriansah et al. 2013). The unexpanded virions at 4 ℃ are covered by 180 copies of the 2D22 Fab (Fig. 2I, 2L), whereas only 120 Fab copies are present on some expanded virions at 37 ℃ (Fig. 2J, 2M), as based on 2D and 3D classification of extracted virus-antibody particles (Fibriansah et al. 2015a). (3) Fusion intermediates. A low pH-triggered rearrangement of the E protein is required for virus–cell membrane fusion during entry of flaviviruses into the cell. The E16 Fab trapped WNV in a prefusion state when the virions were exposed to low pH (Kaufmann et al. 2009). C10, a bNAb for DENV, can structurally lock the E protein of ZIKV at acidic conditions (Rouvinski et al. 2015; Zhang et al. 2016).
-
Many severe human diseases are caused by structurally polymorphic enveloped viruses (e.g., HIV-1, influenza and Ebola viruses). Glycoprotein-specific antibody-inducing epitopes on the viral surface have been studied directly using cryo-ET. The open conformations of the HIV-1 Env spike inducedbyFabb12orCD4/Fab17b have been characterized usingcryo-ETanalysis(Liu et al.2008).Theextenttowhich the C179 antibody bound to the stem domain of hemagglutinin (HA) on the influenza virus was also investigated by cryo-ET, revealing that most of the HA trimers on virions were accessible to this antibody (Harris et al. 2013).
High-resolution SPA of glycoprotein–antibody complexes requires stabilized protein samples, a good example of which is the engineered HIV-1 Env trimer with SOSIP mutations (Sanders et al. 2002). Mature HIV-1 possesses trimers of gp41/gp120 homodimers as the surface spikes. Although some spike epitopes are present on the gp120 monomer, it would be desirable to choose native-like trimers for structural analysis because they are the major target of the neutralizing antibodies elicited by natural infection (Sanders and Moore 2017; Ward and Wilson 2017). SOSIP mutants contain an introduced disulfide (SOS) bond between the gp120 and gp41 ectodomains, and an introduced isoleucine to proline mutation in gp41 to promote trimer formation. Engineered glycoprotein trimers from other enveloped viruses (e.g., Middle East respiratory syndrome coronavirus, MERS-CoV, and parainfluenza virus types 1–4) were designed to present the antigenically optimal prefusion conformation (Pallesen et al. 2017; Stewart-Jones et al. 2018). Glycoproteins from Ebola virus were modified by removing the mucin-like domain, assembled as soluble trimers, and then studied in a complex with the ADI-15878 Fab by SPA (Murin et al. 2018). Except for the above-mentioned trimeric forms, glycoprotein complexes containing more than one viral glycoprotein (e.g., gH/gL/gp42 from Epstein-Barr virus) have also been used for cryo-EM antibody–antigen studies (Snijder et al. 2018).
Cryo-EM structures of glycoprotein–antibody complexes are usually captured in intermediate states. Twenty antibody classes targeting six epitopes on the prefusion closed HIV-1 Env trimer have been characterized using SPA (Table 3) and crystallographic studies (Chuang et al. 2019). 'Breathing' by HIV-1 B41 SOSIP.664 trimers was found to expose the b12 epitope (Ozorowski et al. 2017), and a similar motion by influenza HA protomers was also revealed by SPA (Turner et al. 2019). S230 binding induces fusogenic conformational rearrangements in the SARS-CoV S glycoprotein, while the MERS-CoV S glycoprotein remains its prefusion conformation upon LCA60 binding (Yuan et al. 2017; Walls et al. 2019).
-
VLPs with features that are structurally and immunologically indistinguishable from live viruses are used as alternative models for cryo-EM studies, especially when the viruses need to be manipulated in high-level biosafety facilities. Chikungunya virus (CHIKV) is a mosquitotransmitted human pathogen with T = 4 icosahedral symmetry. On its surface, three E2 molecules form the major component of the viral spike and serve as the main target for antibodies. Because handling live infectious CHIKV requires biosafety level 3 facilities, cryo-EM studies are performed with the safe CHIKV vaccine strain (CHIKV 181/25) (Fox et al. 2015) or CHIK VLPs (Akahata et al. 2010; Sun et al. 2013; Jin et al. 2015). The CHK-265 Fab cross-links two E2 molecules from neighboring spikes (Fox et al. 2015), while the footprints of C9 and IM-CKV063 Fabs on VLPs span the neighboring E2 subunits within one viral spike (Jin et al. 2015). CHK-152 may cross-link the flexible domain B to the domain A within an E2 molecule, and thus inhibiting the exposure of the fusion loop on domain II of E1 (Sun et al. 2013).
VLPs have also served as controllable scaffolds for loading antigenic cargos (Charlton Hume and Lua 2017). Currently, the most commonly used VLPs are rigid icosahedral or helical particles, and flexible platforms are beginning to be promising roles for antigen loading (Hudalla et al. 2014; Rao et al. 2018). To protect against different human papillomavirus (HPV, pseudo T = 7 icosahedral) infections, chimeric VLPs containing the epitopes from three HPV types have been generated and studied by cryo-EM (Li et al. 2018). Recently, computationally designed nanoparticles have also been examined by cryo-EM as a new platform for presenting the respiratory syncytial virus F glycoprotein trimer (Marcandalli et al. 2019). Being able to add antigenic protein modules to tailorable platforms is a tantalizing way of studying highly virulent viruses like Crimean-Congo hemorrhagic fever, Nipah, and Ebola viruses. It is also likely that the cryo-EM characterization of VLPs and nanoparticles will accelerate the development of new vaccine platforms.
-
Cryo-EM has evolved in recent years into a powerful technique for elucidating the structural basis of virusantibody interactions. Compared with traditional X-ray crystallography, cryo-EM offers the following advantages: (1) it can investigate conformational epitopes with sequentially discontinuous residues on icosahedral virions; (2) it avoids tedious screening for diffractable crystals and can be incorporated into a standardized process for rapid and rational vaccine development; (3) it can help with analyzing intrinsic heterogeneous samples, like highly glycosylated viral glycoproteins; and (4) it can be exploited for developing and characterizing high-quality vaccine platforms. Finally, cryo-EM studies of antigen–antibody complexes are beginning to clarify the mechanisms of epitope–paratope recognition at atomic resolution, so we expect that high-resolution cryo-EM structures will play more important roles in future at guiding vaccine development.
-
This work was funded by the National Natural Science Foundation of China (Grant Nos. 31570161 and 31770169), and the "One-Three-Five" Strategic Programs of the Wuhan Institute of Virology, Chinese Academy of Sciences (Grant No. Y605211SA3). We are thankful to the Center for Instrumental Analysis and Metrology of Wuhan Institute of Virology, CAS, for providing cryo-EM support. We also thank Dr. Sandra Cheesman for editing the English text of a draft of this manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
-
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.














 DownLoad:
DownLoad: