HTML
-
The envelope glycoprotein (Env) of human immunodeficiency virus type 1 (HIV-1) plays a key role in viral attachment and entry into target cells through interactions with the CD4 receptor and CXCR4/CCR5 coreceptors (Deng H, et al., 1996; Wyatt R, et al., 1998). Env is the major target for anti-HIV-1 neutralizing antibodies. Although great efforts have been focused on the design of effective HIV vaccines capable of eliciting neutralizing antibodies, it is still a challenge to generate broad and potent neutralization responses. In contrast to the limited success in eliciting neutralizing antibodies, a number of neutralizing antibodies have been isolated from HIV-1-infected donors, which could neutralize most of the circulating HIV-1 strains (Walker L M, et al., 2009; Walker L M, et al., 2011; Wu X, et al., 2010; Zhu Z, et al., 2013) and could protect animals against viral challenge (Hessell A J, et al., 2009; Veselinovic M, et al., 2012). Mapping the epitopes of the neutralizing antibodies and identifying the significant sites that affect neutralization has become a major strategy for HIV-1 vaccine research.
IgG1b12 (b12) was the first broadly neutralizing antibody against HIV-1, which was isolated from a subtype B-infected donor using phage display methods (Burton D R, et al., 1994). Monoclonal antibody b12 can neutralize more than 50% subtype B viral strains and about 30% non-B viral isolates (Binley J M, et al., 2004; Chong H, et al., 2008; Nie J, et al., 2010). In non-human primate challenge models, animals can be protected by b12 with titers as low as 1:5 IC90 (Hessell A J, et al., 2009). In addition, humanized mice can be protected from HIV-1 infection through b12 gene transfer using adeno-associated virus vector (Balazs A B, et al., 2012). The epitope of b12 overlaps CD4 binding sites of HIV-1 Env, which is functionally conserved. Designing immunogens to elicit b12 or b12-like neutralizing antibodies has been attracting great interest since the discovery of this antibody. Although the crystal structure of b12 against core gp120 has been illustrated and the epitope on gp120 recognized by b12 has been clearly defined (Saphire E O, et al., 2001), some key residues influencing b12 neutralization susceptibility, especially those not existing on the contacting surface, have not been clearly determined, which is critical for immunogen design.
CRF07_BC is a major circulating recombinant form (CRF) found throughout China, together with CRF08_BC, CRF01_AE and Thai B, accounting for 95% of reported cases in China (Zhang L, et al., 2004; Zhang Y, et al., 2006). The neutralization susceptibility of CRF07_BC isolates to b12 has been found to vary considerably (Chong H, et al., 2008; Shang H, et al., 2011). Clarification of the mechanisms underlying these variations would be helpful for immunogen design to elicit b12-like neutralizing antibodies. Even within the same subtype, the amino acid sequence diversity of Env can reach as high as 20%(Moore J P, et al., 1996). Consequently, identification of the significant sites that affect neutralization would be challenging. For each chronic HIV-1-infected individual, the in vivo virus population is comprised of millions of strains, which show different genetic and phenotypic characteristics. The identity of the Env sequences from the same individual is significantly higher than those from different donors. If Envs from the same individual show different phenotypic characteristics, it is possible to identify the key amino acid residues which influence these differences. In this study, we found different strains from a CRF07_BC HIV-1-infected individual that showed different neutralization susceptibility to b12. To avoid Taq polymerase-mediated template switching and errors, the use of the single genome amplification (SGA) method and high-fidelity polymerase were employed to amplify full-length env sequences. Based on these sequences, a quasispecies of Env-pseudotyped viruses was constructed to examine the key residues affecting b12 neutralization susceptibility.
-
The HIV-1 gp120 Monoclonal Antibody (IgG1 b12) was obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH from Dr. Dennis Burton and Carlos Barbas (Barbas C F, 3rd, et al., 1992; Burton D R, et al., 1991; Burton D R, et al., 1994; Roben P, et al., 1994), the plasmid pSG3Δenv from Drs. John C. Kappes and Xiaoyun Wu (Wei X, et al., 2002; Wei X, et al., 2003) and the Tzm-bl (JC53-bl) cells from Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc (Derdeyn C A, et al., 2000; Platt E J, et al., 1998; Wei X, et al., 2002). The 293FT cells (Invitrogen, Carlsbad, CA, U.S.A.) were used to prepare the Envpseudotyped viruses by transfection. The plasma sample GX93 was collected from a CRF07_BC HIV-1 infected individual in 2008 in Guangxi Autonomous Region of China. The subtype of the sample was determined by partial env sequences as described previously (Chong H, et al., 2008; Nie J, et al., 2010; Wang S, et al., 2011; Wang Y, et al., 2005).
-
Before RNA extraction, 1 mL of each HIV-positive plasma sample was concentrated to 140 μL by centrifugation at 23,500 g for 1 h at 4 ℃. Plasma RNA was extracted by using a QIAamp viral RNA mini kit (Qiagen, Valencia, CA, U.S.A.). Reverse transcription of RNA to cDNA was performed by using the SuperScript III according to the manufacturer’s instructions (Invitrogen). Eight μL RNA, 1 μL deoxynucleoside triphosphates (10 mmol/L each), and 1 μL Oligo (dT) 20 (50 μmol/ L) were incubated for 5 min at 65 ℃ and then placed on ice for 2 min. First-strand cDNA synthesis was carried out in 20 μL reaction mixtures with 2 μL 10× reverse transcriptase buffer containing 10 mmol/ L dithiothreitol, 5 mmol/L MgCl2, 2 U/μL of RNase inhibitor (RNaseOUT), and 10 U/μL SuperScript III RT. The reaction mixture was incubated at 50 ℃ for 50 min. Following the completion of the reverse transcription step, the reaction mixture was inactivated by being heated to 85 ℃ for 5 min followed by RNase H digestion at 37 ℃ for 20 min (Invitrogen). The resulting cDNA was used immediately for PCR or kept frozen at −20 ℃ until further analysis.
-
The viral cDNA was serially diluted in 96-well plates such that fewer than 28 PCRs yielded an amplification product. According to a Poisson distribution, the cDNA dilution that yields PCR products in no more than 30% of wells contains one amplifiable cDNA template per positive PCR for more than 85% of the time (Butler D M, et al., 2009; Palmer S, et al., 2005). Viral cDNA was then subjected to nested polymerase chain reaction amplification using the following primers: outer sense primer 5’-AGCAAG AAATGGAGCCAGTAGATCC-3’, nucleotides (nt) 5823 to 5847; outer antisense primer 5’-GGTACCT GAGGTCTGACTGGAAAAC-3’, nt 8996 to 9020; inner sense primer 5’-CACCTTAGGCATTTCCTAT GGCAGGAAGAAG-3’, nt 5956 to 5983; and inner antisense primer 5’-GTCTCGAGATACTGCTCCC ACCCCAT-3’, nt 8879 to 8904 (nucleotide position numbering corresponds to HXB2). Final PCR products were cloned into vector pcDNA 3.1D/V5-His-TOPO (Invitrogen), which allows the env genes to be inserted in the correct orientation with a cytomegalovirus promoter for protein expression.
-
Pseudoviruses were prepared by transfecting 293FT cells (6 × 106 cells in 15 mL of growth medium in a T-75 culture flask) with 8 μg of an env/rev expression plasmid and 16 μg of an Env-deficient HIV-1 backbone vector (pSG3ΔEnv) using the Lipofectamine 2000 reagent (Invitrogen). Pseudovirus-containing culture supernatants were harvested 48 hours after transfection, filtered (0.45-μm pore size), and stored at −80 ℃ in 1-mL aliquots until use.
-
We amplified the plasmid template with each forward and reverse primer containing the expected mutations by using PrimeSTAR HS DNA polymerase (Takara, Dalian, China), digested the reaction mixture with Dpn Ⅰ, and transformed it into Top10 chemically-competent cells (Invitrogen). The entire HIV-1 rev/env region was sequenced for the final plasmid preparation (Qiagen). Plasmids containing a single mutation were used as templates to generate double mutations.
-
All the procedures were performed as described previously (Chong H, et al., 2008).
Monoclonal antibodies, plasmid, cells and plasma sample
RNA extraction and cDNA synthesis
Single genome amplification (SGA) and cloning of env DNA Cassettes
Pseudovirus production
Site-directed mutagenesis
DNA sequence, phylogenetic analysis, titration, and neutralization assay
-
We previously observed a very high sensitivity to b12 neutralization in a pseudovirus isolated in an individual (patient GX93) from Guangxi autonomous region in China (Wang S, et al., 2011). To understand whether the extraordinary sensitivity to b12 neutralization is common in the quasispecies from this individual and the mechanism underlying this phenomenon, the SGA method was employed to amplify the full-length env. In total, 24 different env genes were amplified and 24 corresponding Env-pseudotyped viruses were constructed. Phylogenetic analysis of the full-length env nucleotide sequences confirmed that all 24 isolates were grouped within CRF07_BC (Figure 1). The 24 full-length env were clustered into four groups (named as G1, G2, G3 and G4). The amino acid sequence length of the Env proteins was analyzed within and between each group. We found that the length of all the G1 Envs was 852, the Env length of G3 and G4 was 848. For the G2 group, the length of GX93.16 was 852, that of GX93.1, GX93.3 and GX93.10 were 857, and that of GX93.2 was 848.
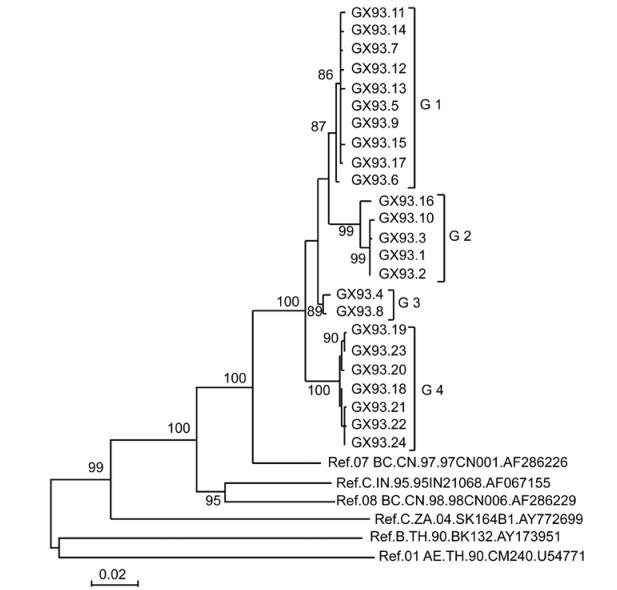
Figure 1. Phylogenetic relationships of HIV-1 env sequences of the quasispecies with reference strains. Horizontal branch lengths are drawn to scale (the scale bar represents 0.02 nucleotide substitution per site), but vertical separation is for clarity only. Values at the node indicate the percentage of bootstraps in which the cluster to the right was found; only values of ≥80% are shown.
-
The 24 Env-pseudotyped viruses cloned from the same individual showed divergent sensitivity to b12 neutralization. According to the sensitivity, the 24 strains were divided into four groups (named as S1, S2, S3 and S4) (Table 1). The ID50 values of the viruses in groups S1, S2, S3 and S4 were > 25 μg/mL, 1 μg/mL–25 μg/mL, 0.1 μg/mL–1 μg/mL, and <0.1 μg/mL, respectively. The groups S1, S2, S3 and S4 contained 4 strains (GX93.1–GX93.4), 3 strains (GX93.5–GX93.7), 10 stains (GX93.8–GX93.17) and 7 strains (GX93.18–GX93.24), respectively. In the virus quasispecies, a large variation in the sensitivity for b12 neutralization was observed, ranging from 0.014 μg/mL for GX93.24 to more than 25 μg/mL for GX93.1. The ID50 difference between the highest and the lowest sensitive strains was more than 1700-fold.
pseudovirus clone length of gp160 Amino acid residues corresponding to HXB2 position ID50 of b12
(μg/mL)Group strain 22 182 276 334 346 386 466 474 698 778 786 S1 GX93.1 857 L V N S R N T N I A G > 25 GX93.2 848 * * * * * * N * * * * > 25 GX93.3 857 * * * * * * * * * * * > 25 GX93.4 848 * * * * * * * * * * * > 25 S2 GX93.5 852 * * * * * * * * * * * 5.356 GX93.6 852 * * * * * * * * * * * 2.859 GX93.7 852 * * * * * * * * * * * 2.255 S3 GX93.8 848 * * * * * * * * * * * 0.734 GX93.9 852 * * * * * * * * * * * 0.454 GX93.10 857 * * * * * * * * * * * 0.325 GX93.11 852 * * * * * * * * * * * 0.280 GX93.12 852 * * * * * * * * * * * 0.258 GX93.13 852 * * * * * * * * * * * 0.230 GX93.14 852 * * * * * * * * * * * 0.188 GX93.15 852 * * * * * * * * * * * 0.172 GX93.16 852 * * * * * * * * * * * 0.167 GX93.17 852 * * * * * * G * V T R 0.117 S4 GX93.18 848 F L S N S D G D V T R 0.092 GX93.19 848 F L D N S D G D V T R 0.080 GX93.20 848 F L S N S D G D V T R 0.040 GX93.21 848 F L S N S D G D V T R 0.020 GX93.22 848 F L S N S D G D V T R 0.015 GX93.23 848 F L D N S D G D V T R 0.015 GX93.24 848 F L S N S D G D V T R 0.014 *Amino acids which were identical to the GX93.1 in the same position are designated with a dot. Table 1. Length of gp160 and amino acid substitutions in Env-pseudotyped virus coinciding with variety of sensitivity to b12 neutralization.
-
To identify amino acid key sites in the gp160 protein that may play a role in the sensitivity to b12 neutralization, full-length Env protein sequences from quasispecies alignment was associated to the ID50 values for b12 neutralization. Eleven amino acid substitutions were found in the most b12-sensitive strains: L22F in the signal peptide, V182L in variable region 2 (V2), N276S in constant region 2 (C2), S334N in C3, R346S in C3,N386D in C3, T/N466G in V5, N474D in C5, I698V in transmembrane (TM) domain, T778A in cytoplasmic tail (CT) and G786R in CT. For the sensitivity groups, all the eleven substitutions were observed in strains belonging to the most sensitive group S4, and were all absent in the resistant groups S1 and S2. For group S3, nine of the ten strains had no such substitutions. Only the most sensitive isolates in this group had four substitutions.
-
To determine whether the identified sites play a role in the variation of the sensitivity to b12 neutralization, two pseudovirus strains were selected as templates to generate the mutations. To verify the function of the identified sites, we made site-directed mutagenesis in two directions, from resistant to sensitive and from sensitive to resistant. The sensitive mutations were introduced into GX93.6 (Figure 2A) and the resistant mutations were introduced into the template GX93.24 (Figure 2B). Eleven mutant viruses were produced for both of the virus strains. From the results, we found that the mutations in 182, 276 and 346 resulted in changes to the sensitivity of b12 neutralization. The L182V mutation decreased the sensitivity of GX92.23 to b12 nearly tenfold (0.143 μg/mL vs 0.015 μg/mL for ID50), while V182L increased the sensitivity of GX93.6 more than twofold (1.339 μg/mL vs 2.853 μg/mL for ID50). The D276N mutation reduced the sensitivity more than twofold in GX93.23 (0.35 μg/mL vs 0.15 μg/mL), while N276D increased the b12 sensitivity by nearly twofold in GX93.6 (1.564 μg/mL vs 2.853 μg/mL). For the mutation in position 346, the serine to arginine decreased the sen-sitivity of GX93.23 more than twofold (0.032 μg/mL vs 0.015 μg/mL), and the reverse mutation in GX93.6 increased the sensitivity more than two fold (1.179 μg/mL vs 2.835 μg/mL).
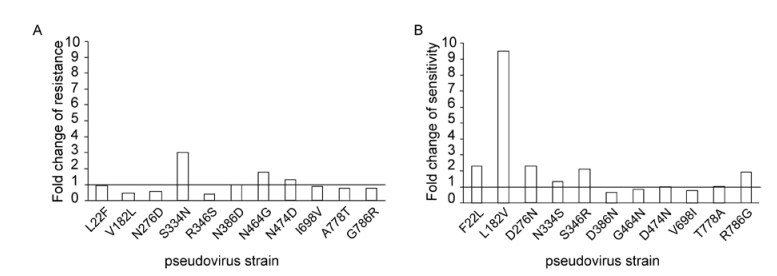
Figure 2. Effect of mutations in gp120 relating to the variation of b12 neutralization sensitivity. Eleven corresponding site-directed mutageneses were introduced into GX93.6 and GX93.23 to produce 11 mutants for each strain. For GX93.6, the aim of the mutagenesis was to enhance the sensitivity of the resistant strain (A). For GX93.23, the aim of the mutagenesis was to reduce the sensitivity of the sensitive strain (B). The results are presented with the (ID50 for mutants)/(ID50 for parental strain).
When the three-site mutageneses were introduced into the same strain, no additive effect was observed. The enhancement of combined three-site mutations was similar to the single V182L mutagenesis. In order to further determine the effect of V182L mutation on b12 neutralization susceptibility, this mutagenesis was introduced to a more resistant strain GX93.2 (ID50 >25 μg/mL). The pseudovirus containing the GX93.2–V182L Env was more sensitive to b12 than the parental pseudovirus, with an ID50 value of 21.35 μg/mL. In further studies using the 18 CRF07/08 BC Envpseudotyped viruses constructed previously (Chong H, et al., 2008), we found that the amino acid residues at position 182 were all valine. The V182L was also introduced to BC17 (ID50 = 13.23 μg/mL) and this produced a twofold enhanced sensitivity to b12, with an ID50 value of 5.85 μg/mL. However, when the V182L mutation was introduced to the CRF01_AE strain GX74.20 (Nie J, et al., 2010), no influence on b12 neutralization susceptibility was observed.
Sequence analysis
Divergent sensitivity to b12 neutralization of the HIV-1 quasispecies obtained from the same patient
Identification of the significant sites in Env protein that related to the sensitivity for b12 neutralization
Site-directed mutagenesis analysis of the identified key sites
-
The key residues of Env, which affect the susceptibility to neutralizing antibody b12, were investigated in this study. Taken from an HIV-1-infected individual, we constructed 24 Env-pseudotyped viruses. The full-length env genes were amplified using SGA methods to avoid Taq-polymerase mediated template switch and were cloned into mammalian expression vectors. Because all the env genes originated from the same individual, the diversity between any two sequences was not greater than 5%. In contrast, great differences were found in the b12 neutralization susceptibility for the 24 pseudoviruses. When the Env sequences and b12 neutralization susceptibility information were analyzed, identification of the key residues became relatively easy. The identified primary residues were subsequently verified through sitedirected mutagenesis.
The epitope on gp120 recognized by b12 is conformation-dependent, and amino acid alterations outside the b12 binding site can influence the sensitivity to b12 neutralization by affecting the conformation of Env. Numerous studies have reported that amino acid substitution accounts for b12 neutralization susceptibility in some viral strains. Pantophlet and colleagues have performed an alanine scanning mutagenesis of a clade B monomeric gp120 and reported numerous alterations that affected b12 binding to gp120; the N276A was reported to increase the affinity of gp120 to b12 (Pantophlet R, et al., 2003). Similar results were observed in our study in which the introduction of N276D to GX93.6 enhanced the sensitivity of the parental strain to b12 nearly twofold. Correspondingly, D276N mutagenesis lowered the neutralization susceptibility of GX93.23 to b12.
The C3 region has been reported to play an important role in affecting b12 neutralization susceptibility of the resistant strains and in the appearance of b12 escape mutants (Duenas-Decamp M J, et al., 2008; Mo H, et al., 1997). Also, the amino acid 346 located in the C3 region moderately regulates the b12 neutralization susceptibility, and therefore, the amino acid position may influence the b12 epitope by affecting the C3 or the overall Env conformation.
The RV144 trial performed in Thailand showed a 31.2% vaccine efficacy for preventing HIV-1 infection (Rerks-Ngarm S, et al., 2009). Antibodies against the HIV-1 envelope variable loops 1 and 2 (V1/V2) domain correlated inversely with infection risk (Haynes B F, et al., 2012). In addition, antibodies directed to the V2 region recognized both a conformational epitope and a linear epitope located at amino acid residues 165–178 in the V2 region (Zolla-Pazner S, et al., 2013), where an isoleucine residue at position 181 (I181) in the V2 region played a crucial role in determining vaccine efficacy (Rolland M, et al., 2012). The V182L mutation identified in our study, when introduced to a resistant strain, could enhance the sensitivity to b12 twofold, while the L182V could increase the resistance to b12 almost tenfold for the b12 sensitive strain.
The proximate amino acid residues at position 181 and 182 may interact with each other to affect the b12-like antibody or the CD4 binding site antibodies. Serum antibodies specific for the CD4bs of Env gp120 have been reported to be responsible for the potent and broad neutralization of HIV-1 strains mediated by broadly reactive sera of some HIV-1-infected patients (Li Y, et al., 2007). Also, the amino acid position 182 is near the C-terminus of V2, where the amino acid sequence is reported to be relatively conserved and believed to affect the interaction of b12 with Env (Gnanakaran S, et al., 2010). The crystal structure of b12 and core gp120 demonstrated that the interaction between them was like fingers fitting into a glove. IgG1 b12 might fit onto gp120 by binding an epitope extending from the V1/V2 loop (Saphire E O, et al., 2001). The amino acid position 182 located in V2 might make the contact between the extending epitope and b12 easier.
Saphire and colleagues have described the crystal structure of b12 interacting with core gp120 and identified 17 residues influencing the affinity of b12 to core gp120 (Saphire E O, et al., 2001). In our study, there were three additional residues identified that were not in the list of those reported by Saphire and colleagues. Of the 17 residues located in the Env protein sequences in our study, no difference was observed between the sensitive and resistant strains. The three additional residues identified in our study were not found on the contacting surface of the crystal structure. These residues may affect the b12 neutralization susceptibility by influencing the conformation of Env protein rather than by direct contacting epitopes.
Identifying vulnerable sites on HIV Env could provide the starting point for a structure-based approach to vaccine design. These sites of vulnerability are often targeted by neutralizing antibodies. Thus, identification of neutralizing antibodies and the elucidation of the epitopes and key residues of these antibodies are powerful tools for immunogen design. By considering the results of the present study, the introduction of amino acid substitution at position 182, 267 and 346 might alter the conformation of Env proteins and help to increase the neutralization susceptibility of virus stains, especially the CRF07_BC strains, to b12. The enhancement of the neutralization susceptibility might predict the exposure of the epitopes for b12. We believe that our results might provide important information to take into account the design and modification of immunogens capable of eliciting b12-like antibody response.
In China, two Phase II clinical trials involving AIDS vaccines are ongoing. The design of these vaccines were based on the CRF08_BC and CRF07_BC HIV-1 strains, and included DNA/MVA (modified vaccinia Ankara) and DNA/Tiantan vaccinia strain (replication-competent)(Kent S J, et al., 2010; Teng T, et al., 2011). Key residues identified from different clades play different roles in the different subtypes (Utachee P, et al., 2014; Utachee P, et al., 2010). We believe that our results might provide important information for recognition of key residues affecting the b12-like antibody susceptibility and help facilitate in vaccine immunogen design for CRF07_BC strains.
-
We would like to thank Dr John C. Kappes and Dr Xiaoyun Wu from the AIDS Research & Reference Reagent Program, National Institutes of Health, United States, for providing pSG3Δenv and TZM-bl (JC53-bl). This study was supported by grants from National Science and Technology Major Project (2012ZX10004701). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
-
All the authors declare that they have no competing interests. All individuals who donated samples in the present study provided informed consent; the study was approved by local ethics committees.
-
YW designed the experiments. JN, JZ, and QC carried out the experiments. JN, WH, and YW analyzed the data. JN and YW wrote the paper. All authors read and approved the final manuscript.













 DownLoad:
DownLoad: