-
Nucleic acid amplification is a valuable molecular tool not only in basic research but also in applicationoriented fields, such as clinical medicine development, infectious diseases diagnosis、gene cloning and industrial quality control etc. Several amplification methods have been developed already, such as polymerase chain reaction (PCR) (19), self-sustained sequence replication (3SR) (6), nucleic acid sequencebased amplification (NASBA) (2), strand displacement amplification (SDA) (22) and rolling circle amplification (RCA) (14) etc. Most of these methods can amplify targets by many magnitudes of order using its own particular mechanism to re-initiate new rounds of DNA synthesis, but still have many shortcomings, including the requirement of precision instruments and elaborate methods for product detection. LAMP is a novel nucleic acid amplification method developed by Notomi et al, which amplifies DNA with high specificity, sensitivity and rapidity under isothermal condition using a set of four specially designed primers and a Bst DNA polymerase (17). In this article, we overview the current status of LAMP and recent developments associated with the method.
HTML
-
Loop-mediated isothermal amplification(LAMP)is performed in a microtube using a regular laboratory water bath, which is kept at 65℃ for 60 min and then heated at 80 ℃ for 2 min to end the process. The reaction is carried out in a volume containing 1× LAMP Buffer, MgSD4, betaine, dNTPs, DNA template, a set of four specially designed primers and Bst DNA polymerase. The amplification mechanism is illustrated in Fig. 1.
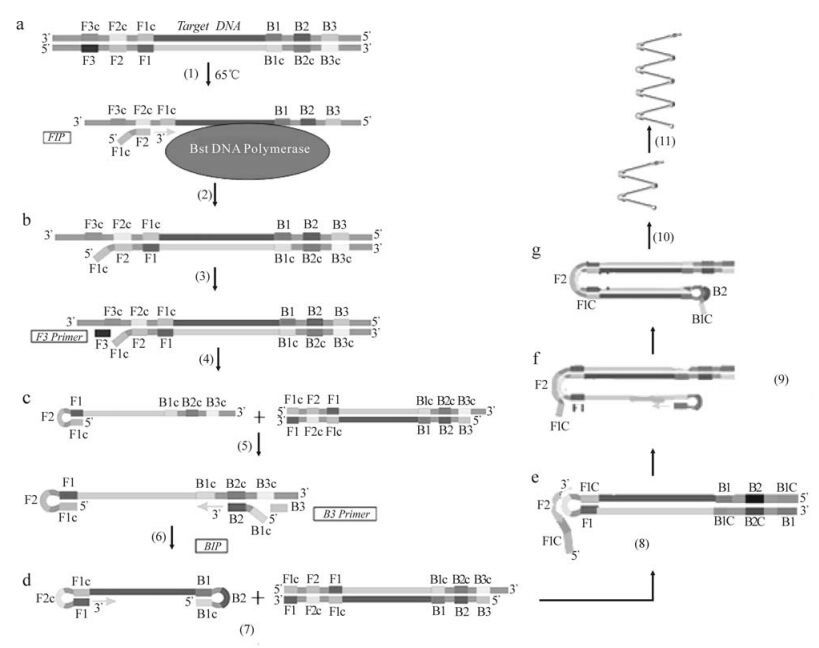
Figure 1. Schematic representation of the LAMP mechanism (This photograph comes from http://loopamp. eiken.co.jp/e/lamp/principle.html with minor modification).
The amplification is composed of two main steps—starting material production and cycling amplification (17). In the first step, the inner primer FIP first hybridizes to F2c to initiate the strand synthesis and produces a DNA strand complementary to the template (structure b) at the temperature around 65℃. Then, the out primer F3 slowly hybridizes to F3c to initiate the strand displacement synthesis under the strand displacement activity of the Bst DNA polymerase, producing a FIP-linked complementary strand, which can form a looped out structure at one end (structure c). This structure serves as template for BIP-initiated strand synthesis and subsequent B3-primed strand displacement synthesis, generating a dumb-bell form (structure d). This form of DNA then serves as the starting material for LAMP cycling amplification. In the second step, first, the dumb-bell form DNA structure is quickly converted into a stemloop DNA by self-primed DNA synthesis (structure e). Then, FIP hybridizes to the loop of the dumb-bell DNA and primes strand displacement DNA synthesis, releasing a single strand DNA which quickly forms a stem-loop structure at the 3' end because of complementary B1c and B1 regions, finally, generating a gapped stem-loop DNA (structure f). Subsequent self-primed strand displacement synthesis yields one repaired stem-loop DNA (structure g) with a stem elongated to twice the length and a loop at the opposite end. The product then serves as template for a BIP-primed strand displacement reaction in the subsequent cycle amplification until the reaction is ended and the final products are a mixture of stem-loop DNAs with various stem lengths.
-
LAMP is highly specific for amplification of the target DNA sequence. This is most probably attributable to recognition of the target by six / eight independent sequences in the initial reaction stage and by four/six independent sequences during the second stage. Imai et al. used LAMP to investigate eight H5-influenza virus samples and twenty-two other influenza virus samples and found that the H5-specific primer set amplified only the H5-influenza virus while nonspecific amplification products were not observed.
-
LAMP amplifies DNA with high sensitivity with the detection limit of few copies, comparable to that of PCR. Dukes et al. found that in the detection of foot-and-mouth disease virus (FMDV), LAMP was even more sensitive than the TaqMan® real-time PCR (3). In our study on pseudorabies virus (PRV) detection by LAMP, 10 fg DNA sample could be detected, which was 100~1000-fold higher than that of PCR.
-
The products are a mixture of stem-loop DNAs with various stem sizes which would enable subsequent simple and selective detection of products. Agarose gel electrophoresis detection, turbidity detection and fluorescent detection are often used in LAMP, while the latter two have the potential to be applied in field practice.
-
LAMP is simple and easy to perform once the special primers are designed, requiring only a Bst DNA polymerase and a regular laboratory water bath for reaction.
-
Once a suitable reaction system is established, only one hour is required to amplify the target DNA sequence by more than 109-fold.
It is anticipated that with the advantages of specificity, sensitivity, rapidity and easy manipulation, LAMP will be a powerful molecular tool for the DNA amplification and can be used widely in practice in the field.
High specificity
High sensitivity
Easy detection
Simple manipulation
High efficiency
-
Since the development of LAMP in 2000, many modifications and improvements have been reported.
LAMP product was previously detected by agarose gel electrophoresis and visualized by staining with ethidium bromide, which is either time consuming and does not meet the requirement of field tests. However, it was observed that a large amount of white precipitate was produced during the LAMP amplification (Fig. 2). Accordingly, Mori et al. clarified the cause of the precipitate production and used this to establish a new method for detection of LAMP products (15). Their method is based on the observation that the increase of the precipitate correlates with the amount of DNA synthesized and real-time monitoring of the LAMP reaction was achieved by measurement of turbidity.
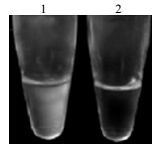
Figure 2. White precipitate preduced in LAMP reaction. 1. Negtive control; 2. Positive LAMP reaction.
Nagamine et al. developed a method that could accelerate the LAMP reaction by using additional primers, termed loop primers (18). Loop primers hybridize to the stem-loops, with the exception of the loops that are hybridized by the inner primers, and prime strand displacement DNA synthesis. They found that this method could halve the reaction time to facilitated rapid application of the LAMP process. In addition, amplification could occur from 103 copies of the target using the loop primers while under the absence of loop primers, the amplification required more than 104copies, which suggested that LAMP reaction with the loop primers provided higher sensitivity and efficiency.
Many reports have described LAMP assay as being less affected in terms of sensitivity by the presence of inhibitory substances than PCR. Kaneko et al. have performed a detailed evaluation of the tolerance of LAMP against a culture medium and certain biological substances (11). They used the herpes simplex virus type (HSV) diluted with Minimum Essential Medium (MEM) as sample material. The results showed that HSV DNA was detected at a 10-5 dilution in both samples with and without DNA extraction, indicating that MEM did not affect the sensitivity of LAMP. Other substances were all tested including PBS, Serum, Plasma, Urine, and Vitreous (Table 1), which showed that the tolerance of LAMP for biological substances was superior than PCR, suggesting the DNA extraction step could be omitted in a LAMP assay. This would save time, labor and cost, which would allow LAMP assay to be developed as a superior diagnostic method for the field test.

Table 1. Effects of biological substances on LAMP and PCR assay
-
LAMP has already been widely used in life science, especially in detection of microorganisms, infectious diseases diagnosis and embryo sex identification. Here, we summarized some of the most important applications.
-
LAMP has already been used for bacteria detection. Aoi et al. used LAMP as a simple method for monitoring ammonia-oxidizing bacteria (1). Their results showed that it was possible to quantify the initial target DNA with sensitivity down to 102 copies while the background DNA from non-targeted bacteria did not affect the quantitative capability of LAMP, which suggested that the LAMP is effective for monitoring bacteria and their gene expression in environment.
-
Some species of Brettanomyces could cause turbidity or off-flavors in wines, beer and soft drinks, resulting in significant economic losses worldwide. Hayashi et al. established an identification method using LAMP which is advantageous in terms of specificity, sensitivity, rapidity and simple operation compared with PCR (8). They designed primer sets specially targeting the ITS region of Brettanomyces (Dekkera anomala, Dekkera bruxellensis, Dekkera custersiana and Brettanomyces naardenensis) which could detect 1×10 CFU/mL of Brettanomyces yeast isolates from water, beer, wine and soft drinks.
-
RT-PCR with H5-specific primers recommended by WHO has been used worldwide for H5-influenza virus detection following the outbreak of highly pathogenic avian influenza (HPAI) caused by the H5N1 influenza virus in the end of 2003. However, this assay is timeconsuming and requires precision instruments which prevent its being widely used, especially in some poor countries. Recently, Imai et al. have used LAMP to develop a rapid laboratory diagnostic system for HPAI and the sensitivity and specificity of the test have been validated (10). The results suggested that the present LAMP system is a useful diagnostic tool for the HPAI-H5N1 virus.
-
Predetermination of embryo sex is very important for the embryonic research. Detection of Y chromosome-specific sequences has widely been used to predict the sex of offspring. Hirayama et al. have established an efficient procedure for water buffalo embryo sex determination by identifying a Y chromosome-specific sequence using LAMP, while a 12S rRNA was also amplified by LAMP as a control reaction in both male and female (7). The results showed that the minimal amount of the template DNA required appeared to be 0.1-10 pg, and the total time for embryo sexing determination, including DNA extraction was about 1 h.
-
LAMP has also been successfully applied in diagnosis of other disease viruses, such as detection of Hemorrhagic septicaemia virus (VHSV) (20), Cytomegalovirus (CMV) (21), Ebola virus(EBOV) (13), Epstein-Barr virus(EBV) (9), Fish iridovirus (16), Human herpesvirus 8 (12), infectious hypodermal and hematopoietic necrosis virus (IHHNV) (23), tomato spotted wilt virus (5), Tomato yellow leaf curl virus (4). In current work, we are planning to apply the LAMP method to detect the important pig infectious disease viruses, such as PRV and FMDV, to help strengthening virus surveillance.
Detection of ammonia-oxidizing bacteria
Detection of Brettanomyces
Detection of H5N1 avian influenza virus
Embryo sexing identification
Other Applications
-
In conclusion, LAMP is a novel method of nucleotides amplification which can amplify a few copies of DNA in less than an hour under isothermal condition and with great specificity. There have been many reported researches which have proven this method to be a successful molecular tool for the amplification of target DNA sequence. The method provides us with a powerful biological tool to be used in the research of clinical medicine, diagnosis of infectious diseases, genetic disorders and genetic traits.














 DownLoad:
DownLoad: