-
Influenza infection occurs in as much as 5%-15% of the world population, resulting in 3-5 million cases of severe illness and up to 500 000 deaths annually. In the U.S. alone, the flu epidemics lead to approximately 300 000 influenza-related hospital admissions and 36 000 influenza-related deaths annually in addition to an estimated cost of $12 billion per year[7, 10]. The current seasonal influenza vaccines are produced using the strains recommended by the World Health Organization about 6-8 months ahead of the targeted season[2]. These vaccines typically contain two sub-type strains of influenza A virus and one influenza B stain, which are predicted to be the most likely strains to circulate in the upcoming year.
At present, the viruses for production of the inactivated vaccine are propagated in the chicken embryo. Although the purification process is effective, the protein of the chicken embryo can't be removed completely and may cause allergic reactions in vaccinated populations. Another disadvantage is that the serial passage in chicken embryo could cause a change in HA antigenicity. Such changes result into a mismatch between the circulating strains and the vaccine components. Additionally, the growth of influenza viruses in eggs often selects variants that differ in their glycosylation patterns from the original clinical isolates. Specifically and most importantly; the insufficient supply of embryonated eggs could limit the production of vaccines when the pandemic influenza occurs.
The use of different cell lines for vaccine virus growth could largely overcome these problems, provided that the yields were satisfactory and safety issues could be addressed[1]. Although Vero cells have already been used for producing many kinds of human vaccines and its safety is generally accepted, influenza virus can't always adapt to Vero cells. In this study, we successfully screened an influenza B virus strain which grows well in Vero cells, suggesting that it is possible to prepare the influenza vaccine using Vero cells as substrate[11].
HTML
-
Vero cell line were obtained from the American Type Culture Collection (ATCC) and conserved in the Institute of Medical Biology, Kunming. The influenza B viruses in this study (Table 1) were isolated from the patients with an fever over 38℃ and diagnosed as influenza virus infection.

Table 1. The HAT of twelve Strains from Clinical patients after propagation in Vero cells
-
Samples from patients infected with influenza B viruses were collected at the Jingping People's Hospital of Yunnan Province. Vero cells were infected with the throat and nose swab material, and incubated at 33℃ until the appearance of cytopathic effect (CPE), the viruses were harvested and used for the later study.
-
Viruses were propagated in Vero cells in DMEM/ F12 medium containing 1μg/mL of trypsin at 33℃. The viruses were harvested at 72 h post-infection. The hemagglutination assay was performed to measure the content of viruses with 1% suspension of chicken red blood cells for 30 min, and the results were indicated by HAT (Hemagglutination titer) units.
-
HAI (hemagglutination inhibition) test: The reagents for hemagglutination inhibition test were obtained from the Chinese National Influenza Center. The anti-serum of H1, H3, BVintoria, and BYamagata (2-fold serial dilution) were mixed with the viruses (containing 4 HAT units) at 33℃ for 30min respectively. Then the hemagglutination assay was carried out with 1% suspension of chicken red blood cells for 30 min.
RT-PCR: The RNeasy Kit (Qiagen) was used to extract vRNA from 100 μL of virus. The RNA was eluted into 40μL of H2O. For RT-PCR of the HA1 segment, the One Step RT-PCR kit (Qiagen) was used according to the protocol provided. 20 μL of RNA was used for each reaction. The RT reaction was performed for 50 min at 50℃, followed by 15 min at 94℃. PCR reaction was performed by using the B type upstream primer B-HA1-f: CGAATCTGCACTG GGATAACATC, and the B type downstream primer B-HA1-r: TGCACCATGTAATCAACAACAA. At the same time, PCR reaction were performed by using the A type upstream primer A-HA1-f: CTCGAGAGC AAAAGCAGGGG, and the A type downstream primer A-HA1-r: CTGCTATTGCGCTGAATAG [8]. The PCR mixtures were treated for 4 minutes at 94℃ and was followed by 30 cycles of amplification: denaturation at 94℃ for 30 sec, renaturation at 55℃ for 45 sec, and extending at 68℃ for 90 sec.
-
The HA1 gene fragment was sequenced by the Takara company (Dalian). The results were analyzed using MEGA 4.0. The cladogram was drawn by neighbor joining method. Genetic distances were calculated by the Kimura two-parameter model.
Cells and Viruses
Primary isolation
Propagation of viruses in Vero cells
Type Identification
Genetic analysis
-
Twelve strains of influenza B virus were inoculated into Vero cells. Most of them could not be propagated in Vero cells, and the HAT titer decreased to an undetectable level after 3 generations. However, the B/Yunnan/2/2005va (B) strain, although the HAT titer are lower at the first few passages, could proliferate efficiently in Vero cells, with the hemagglutination titer (HAT) upto 1:512 after being propagated 9 generations in Vero cells (Table 1).
Furthermore, the B/Yunnan/2/2005va (B) virus was propagated consecutively to the 21st generation in Vero cells, and the HAT maintained between 1:512 to 1:1024. The HAT was 1:768 even at the 21st generation.
-
HAI test: The results of HAI test showed that the HAI titer of H1 and H3 was at 1:2; the HAI titer of BY was at 1:512 and the HAI titer of BV was at 1:16. According to this data and the introduction of the kit, we can determine the types or lineages of the influenza virus if there were 4 differences between the two HAI titers. So the B/Yunnan/2/2005va (B) could be classified as the Yamagata lineage of influenza B virus.
Identification of genotype: After PCR amplification, a 750bps band was observed on the 1% agarose gel in the PCR products of the B type primers, while not in the products using the A type primers (Fig. 1). The results further proved that B/Yunnan/2/2005va (B) strain belongs to the influenza B virus.
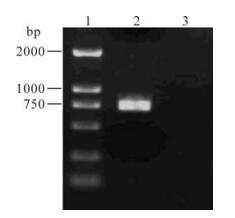
Figure 1. RT-PCR for amplification of the HA1 segments. RT-PCR was performed with the B and A type primers B-HA1-f/B-HA1-r and A-HA1-f/A-HA1-r. The PCR products were subjected to gel electrophoresis on a 1% agarose gel. Lane 1, DNA ladder; 2, PCR products of the B type primers; 3, PCR products of the A type primers.
-
Our data included the HA1 gene segments of influenza B viruses from GenBank. The numbers are summarized in Table 2. MEGA 4.0 was used to construct the HA1gene tree (Fig. 2).

Table 2. The GenBank number of these virus strains in the gene tree.
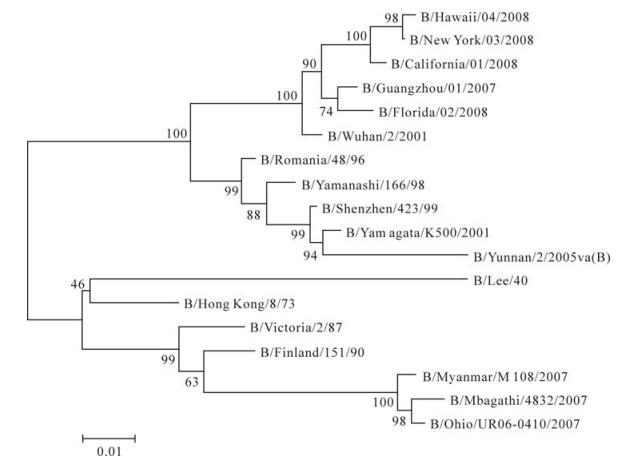
Figure 2. Phylogenetic analysis of the B/Yunnan/2/2005va (B) by the neighbor-joining method. Genetic distances were calculated by the Kimura two-parameter model. The scale indicates the relative phylogenetic distance.
Phylogenetic analysis showed that B/Yunnan/2/ 2005va (B) shared 97% nucleotide sequence similarity with the HA1 gene from the B/Yamagata/K500/2001 strain. The cladogram was constructed using the neighborhood joining method. Genetic distances were calculated by the Kimura two-parameter model. The scale indicates the relative phylogenetic distance. Among these strains of influenza B virus, the genetic distance was in a range from 3% (comparison of B/Yunnan/2/ 2005va (B) with B/Yamagata/K500/2001 and B/Shenzhen/423/99) to 20% (comparison of B/Yunnan/2/2005va (B) with B/Mbagathi/4832/2007 and B/Lee/ 40). The genotypes of the HA1 fragment of the B/ Yunnan/2/2005va (B) strain was determined mainly based on the basic differences among nucleotide sequences on the tree. Thus, the B/Yunnan/2/2005va (B) was classified as belonging to the Yamagata lineage of influenza B virus.
Adaptation of viruses to Vero cells
Characterization of the virus
Phylogenetic dendrograms of the B/Yunnan/2/ 2005va (B) strain
-
Investigation of live influenza vaccines in Russia and USA over the past 30 years indicated that these vaccines appeared to be safe and genetically stable, but not always immunogenic. Improvements of the immunogenicity of the live vaccine could be achieved by producing of the vaccine strains in mammalian cell lines. Adopting this approach could avoid mutations and changes in the receptor binding sites along with the antigenic sites of viruses that result from cultivation in embryonated hen eggs. It is important that the antigenicity and glycosylation pattern of vaccine strains are identical to human viruses clinically isolated from swabs. It is well known that viruses grown in human cells can maintain these characteristics; Therefore, vaccines derived from these cells can be more immunogenic and can match the actual circulating virus strains. Moreover, production of the vaccine in the Vero cell line can eliminates the risk of allergic sensitization and allergic reactions to ovalbumin and other hen's egg proteins.
As mentioned in other studies, the infectivity of influenza viruses will decrease when propagated in cells. We found that the HAT (hemagglutination titer) of the virus isolated and passaged in Vero cells decreased from the second generation[12]. Under selection pressure, only the infective virus particles can survive and proliferate in Vero cells. Although the defective viruses could also agglutinate chicken erythrocyte, the HAT appeared to be decreasing during passage in Vero cells. This is because this virus could not penetrate into and propagate in Vero cells, so the concentration of the virus was diluted by the culture medium. In our study, we screened of the B/Yunnan/2/2005va (B) strain, which appeared to have high HAT after being passaged after 7 times in Vero cells. The virus showed low HAT at early generations, suggesting that the defective virus particles would interfere with the infection virus particles when replicating in the cells. According to the principle of species evolution, if the influenza B virus was passaged over two generations without decreasing of HAT, it could be considered that the virus is adapted to the cells [3, 1].
Phylogenetic analysis showed that B/Yunnan/2/ 2005va (B) strain shared 97% nucleotide sequence of the HA1 gene with the B/Yamagata/K500/2001 strain. The genetic distance between B/Yunnan/2/2005va (B) with other strains was in a range from 3% to 20%, thus, the B/Yunnan/2/2005va (B) virus was classified as belonging to the Yamagata lineage of the influenza B virus [5, 8]. The same conclusion was drawn in the HAI test and the RT-PCR result.
The successful adaptation of this B virus strains to Vero cells is important for influenza vaccine development using Vero cell as substrate. However, the problem of adaptation may still exist for other B virus strains recommended for annual vaccine production. This could be overcame by the application of the reassortant technique or reverse genetics, both of which are now used to prepare the influenza virus strains for vaccine production [4, 5]














 DownLoad:
DownLoad: