-
Human papilloma virus (HPV) is a double-stranded DNA virus approximately 8 kb in size. To date, more than 100 HPV types have been identified [3, 12], of which about 30 are associated with genital infections [2, 4]. Although most HPV infections are transient, persistent ones can lead to genital warts, intraepithelial neoplasia and genital cancer. Genital warts are the most common sexually transmitted infection and mainly affect younger people. The HPV prevalence rate in genital warts has been estimated at 98.2% and the most common types are HPV-6 or 11[11]. The incidence of genital warts is high and appears to be increasing[19]. Early vaccination can provide the greatest chance of preventing genital warts. Merck's HPV Quadrivalent (Types 6, 11, 16, and 18) vaccine, for example, can prevent infection with HPV 6 and 11, which are also the most common types identified in patients with genital warts in China. Nevertheless, the positive rate and the HPV prevalence may vary between different regions.
The neutralizing antibody titers induced by natural infections, especially in patients with genital warts, are often low[10, 11, 16], which may be due to the deficiency of an effective and convenient detection method. In a previous report, the positive rate of neutralizing antibodies in patients with condyloma acuminata was 25% (5/20); whereas the presence of HPV neutralizing antibodies in the serum of human patients with natural HPV infection has yet to be verified. Therefore, to analyze the relationship between neutralizing antibodies and HPV DNA types in women with genital warts, we tested the type-specific neutralizing antibodies in sera and HPV types in corresponding cervical swabs.
Analysis of HPV genomes have been reported for a long time. However, the most published genotype is HPV-16, with 17 complete genomes of different HPV-16 isolates identified, whereas research on other HPV subtypes is limited. Therefore, it is difficult to compare genome homology of different isolates. In our assay, we aimed to develop a PCR method for amplification of the full HPV sequence from HPV DNA positive samples and analyze the homology between the sequence we obtained and the reported genome, so as to establish a PCR method that will be useful for future sequence analyses.
HTML
-
In total, we collected samples from 50 patients with genital warts between Aug. 2007 and Dec. 2007, who were living in Tianjin city, China. Each patient provided a serum sample and corresponding cervical swab, the average age of the population was 31.3 (±7.4) years. All patients provided informed consent.
Sera positive for HPV-16 and HPV-18 type-specific neutralizing antibodies from goats vaccinated with HPV vaccine were kindly provided by Prof. Ningshao Xia from Xiamen University.
Plasmids (p16LLw, p18LLw, p58LLw, p45LLw, p6LLw, p11L1w and p11L2w) encoding the structural genes L1 and L2 were obtained from Dr. John T. Schiller (National Cancer Institute, Bethesda, MD).
In the study, the reporter plasmid pcDNA3.1-hGFP was constructed by inserting a human modified green fluorescent protein (GFP) reporter gene which has 100% identity with the published sequence DQ768212 into the pcDNA3.1 vector (Invitrogen, Carlsbad, CA). The GFP gene was amplified from the plasmid pDRVI 1.0-EGFP, which was given by Prof. Jianqing Xu (Chinese Center for Disease Control and Prevention). The size of the reporter plasmid was 6 148 bp.
-
Production of pseudoviruses using 293FT cells (Invitrogen) was accomplished using previously reported methods[5] and the titers of the pseudo-viruses were determined by TCID50 assays. The inoculum sizes of 200-300 TCID50/50 μL were chosen for all the pseudovirus types (HPV-16, -18, -58, -45, -6 and -11) in the neutralization assays. Serial dilutions of the sera to be tested were prepared with ranges of dilution of 1:40-1:10 240. Diluted sera (60 μL) and pseudoviruses (60 μL) were mixed in a dilution plate and placed on ice for 1 h. To monitor inter-assay variability, there were at least two control wells that contained cells plus 120 μL of medium instead of a pseudovirus and serum, and these wells acted as blank samples to detect and deduct the background fluorescence. Four wells of pseudovirus-infected cells that contained 60 μL of medium instead of a neutralizing serum were also evaluated, and these wells acted as negative controls to calculate the infection inhibition rate of the wells containing the neutralizing serum. Two wells of diluted positive sera were used as positive control in each run. Next, 100 μL of each mixture was added to cells which had been pre-plated into 96-well cell culture plates at 1.5×104 cells/100 μL/well before 6-8 h. After incubation for 72 h, the cells were trypsinized and resuspended in DPBS (Dulbecco's phosphate-buffered saline) (Invitrogen) containing 1% FCS, and the percentage of positive cells was determined by FACS analysis. The titer in each serum sample was defined as the reciprocal of the highest dilution that reduced the rate of transduced cells by at least 50% compared with wells containing a pseudovirus but no antibody. With regard to the serum samples, titers of ≤40 were considered negative, while titers of > 40 were considered positive [18].
-
DNA Preparation. Each cervical swab was placed in a tube containing 400 μL PBS (phosphate-buffered saline). After 1 h, the swab was removed and total DNA was extracted from the cell suspension using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Finally, 200 μL of DNA solution was generated and 2 μL of this DNA solution underwent PCR amplification for DNA genotyping.
HPV DNA Genotyping. To identify and specify the HPV types contained in each sample, HPV genoty-ping was performed using a HPV GenoArray DNA Test kit (Hybribio, Hong Kong, China). A total of 21 types of HPV, including 13 high-risk types (HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59 and -68), five low-risk types (HPV-6, -11, -42, -43 and -44) and three undetermined risk types (HPV-53, -66 and -CP8304), could be genotyped. In this assay, HPV DNA was amplified with HPV-specific primers and evaluated by Hybribio's proprietary Flow-through Hybridization Technology, which can discriminate between HPV genotypes.
-
Primer Design. After the HPV DNA genotypes were determined, primers were selected from both ends of each type-specific full-length genome. The primer sequences are described in Table 1.

Table 1. Oligonucleotide sequences used for amplification of full-length HPV genomes
PCR Procedures: PCR amplification was performed in a final volume of 25 μL. Each PCR mixture contained 1.25 U of LA Taq (TaKaRa, Tokyo, Japan), 10×LA PCR Buffer II (Mg2+ Plus), 400 μmol/L of each deoxynucleotide triphosphate, 0.4 μmol/L of each primer and 2 μL of DNA template extracted from a swab. All amplifications were performed with the following cycling profile: 94℃ for 1 min; 10 cycles of 30 s at 98℃ and 8 min at 68℃; and 20 cycles of 30 s at 98℃ and 8 min at 68℃ in the first cycle, with 20 s added to each cycle in the subsequent cycles. The PCR products were purified with a QIAquick Gel Extraction Kit (Qiagen) according to the manufacturer's instructions, cloned into the pGEM-Teasy clone vector (Promega, Madison, WI) and sequenced.
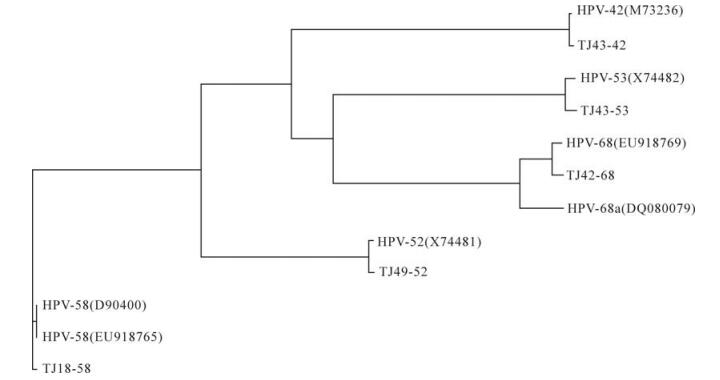
Homology Analysis: The homologies of the entire sequences of the obtained clones and the genomes published in GenBank were analyzed by BLASTing in NCBI. The identities of the corresponding ORFs were aligned using the Vector NTI software (Version 6.00; InforMax Inc., Frederick, MD) for both the nucleotide and amino acid sequences. Vector NTI was also used to generate a phylogenetic tree of the obtained genomes and their corresponding published sequences as follows: HPV-42 (M73236); HPV-52 (X74481); HPV-53 (X74482); HPV-58 (D90400, EU918765) and HPV-68 (DQ080079, EU918769). The partial sequence of human cellular DNA/human papillomavirus proviral DNA (M73258) was also used. The accession numbers of the sequences TJ43-42, TJ49-52, TJ43-53, TJ18-58, and TJ42-68 were GQ472847, GQ472848, GQ472849, GQ472850, GQ472851.
Samples, Positive Sera and Plasmids
Detection of Neutralizing Antibodies using GFP Pseudovirus-based Neutralization Assays
HPV DNA Genotyping
Amplification of The Complete HPV Genome and Sequence Analysis
-
The sera obtained from patients with genital warts were tested for neutralizing antibody using six types of pseudovirus neutralization assay, for HPV types 16, 18, 58, 45, 6 and 11. Overall, 18 of the 50 (36.0%) sera samples were seropositive and all six antibody types tested were detectable in one or more sample. 11 of the 18 positive cases were positive for only one type, including three cases, which were positive for HPV-16 with titers ranging 160-640. Eight cases were positive for HPV-6 with titers ranging 160-2 560. Four of the other seven positive cases were neutrali-zation antibody positive for two types, HPV-18 and -11 (titers of 160 for both types), HPV-18 and -45 (titers 160 for both types), HPV-16 and -6 (titers of 160 for both types), and HPV-18 (titer 640) and -6 (titer 160). The remaining three positive cases were positive for three types each; one case was positive for HPV-16, -18, and -6 (titers of 160 for all types), one was positive for HPV-58, -45, and -6 (titers of 160 for all types), and the third was positive for HPV-16, -18(titers of 640 for both) and -58 (titer of 160) (Table 2).

Table 2. The detection rates of HPV DNA and neutralizing antibodies (NAb)
-
Thirty of the 50 (60.0%) DNA samples were HPV DNA positive. 23 of the 30 (76.7%) positive cases were positive for one genotype, of which five (21.7%), four (17.4%), two (8.7%), two (8.7%), two (8.7%), two (8.7%) and two (8.7%) were positive for HPV-68, -58, -6, -33, -16, -18, and -52, respectively, and only one case was positive for each of the other types (HPV -53, -39, -11, -56). Seven of the 30 (23.3%) positive cases were detected for multiple infections. Two of the seven cases were infected with two genotypes, one patient with HPV-16 and HPV-33, and the other one with HPV-16 and HPV-68. Three of the seven cases were infected with there genotypes, one case was infected with HPV-33, -59 and -CP8304, one case was infected with HPV-11, -68 and -18, and the other case was infected with HPV-11, -68 and -16. Five types were detected in both of the remaining two multiple infection samples, HPV-16, -31, -52, -58, -68 for one sample and HPV-16, -58, -53, -6, -42 for the other sample. In total, 15 HPV genotypes were detected, including 10 high-risk types. The positive rates were 18% for HPV-68, 14% for HPV-16, 12% for HPV-58, 8% for HPV-33, 6% for HPV-18 and HPV-52, and 2% for HPV-31, HPV-39, HPV-56 and HPV-59. For three low-risk types, the positive rates were 6% for HPV-6, 6% for HPV-11 and 2% for HPV-42. For the remaining two undetermined risk types, the positive rates were 4% for HPV-53 and 2% for HPV-CP8304) (Table 2).
-
Neutralizing antibodies against six HPV types (HPV-16, -18, -58, -45, -6 and -11) were detected in the present study. Thus, the associations between HPV DNA and neutralizing antibodies were analyzed, based on these six HPV types (Table 3). In total, 30 of the 50 samples were positive for one or more of the six types of HPV DNA, while only 18 of the 50 samples were positive for the corresponding neutralizing antibodies. The concordance between HPV DNA and the corresponding neutralizing antibodies was 56%. Of the 50 cases, 26% (13/50) were positive for both HPV DNA and the corresponding neutralizing antibodies, 10% (5/50) were positive for neutralizing antibodies and negative for the corresponding HPV DNA, 34% (17/50) were negative for neutralizing antibodies and positive for the corresponding HPV DNA, and 30% (15/50) were negative for both neutralizing antibodies and the corresponding HPV DNA. The geometric means of the neutralizing titers were 303 and 485 for the DNA-positive and -negative samples, respectively.

Table 3. Associations between HPV DNA and neutralizing antibodies
Of the cases positive for neutralizing antibodies, 12 of the 18 (66.7%) patients were positive for HPV-6, which was the most prevalent type detected in the neutralization assay, so the type specific analysis was performed for HPV-6. The concordance between HPV-6 DNA and its type-specific neutralizing antibodies was 78%, which was statistically significant (X2=7.36, P < 0.05) (Table 4). The geometric means of the HPV-6 neutralizing titers were 640 and 320 in the HPV-6 DNA-positive and -negative samples, respectively.

Table 4. Associations between HPV-6 DNA and neutralizing antibodies
-
Only five isolates (TJ43-42, TJ49-52, TJ43-53, TJ18-58 and TJ42-68) of full-length HPV genomes were successfully amplified with type-specific primers. Sequence analyses showed that these five isolates comprised 7 921, 7 960, 7 863, 7 824 and 7 830 bp, respectively, and that all of them contained eight ORFs. The full-length nucleotide sequences of isolates TJ43-42, TJ49-52, TJ43-53, TJ18-58 and TJ42-68 showed 99.3%, 98.7%, 98.3%, 99.4% and 92.9% identities with HPV-42, -52, -53, -58 and -68a, respectively, as published in GenBank. The identities of the corresponding L1 sequence were 99.4%, 99.2%, 98.3%, 99.6% and 92.4%, while the TJ42-68 L1 sequence had 98.4% identity with an incomplete sequence (6 042 bp) of HPV-68b (ME180-HPV) L1. Thus, isolates TJ43-42, TJ49-52, TJ43-53 and TJ18-58 were much more conserved compared with their corresponding types, while isolate TJ42-68 may belong to HPV-68b. The phylogenetic tree of the genomes obtained in this study and the corresponding published types in GenBank confirmed the results of the sequence analyses (Fig. 1).
Detection of Neutralizing Antibodies in Sera
HPV DNA Typing
Association between HPV DNA and Neutralizing Antibodies
Amplification and Sequence Analyses of the Complete HPV Genomes
-
Most neutralization assays available are cumbersome or lack sufficient sensitivity to detect neutralizing antibodies in response to natural HPV infection, which does not induce a strong antibody response. To assess the production of neutralizing antibodies at such low levels, a more sensitive approach using a pseudovirus-based neutralization assay was developed and was found to be useful for quantifying potential type-specific protective antibody levels in HPV [5, 18]. We applied the GFP pseudovirus-based neutrali-zation assay to detect the type-specific neutralizing antibodies of 50 serum samples obtained from patients with genital warts in Tianjin city. The neutralizing antibody positive rate was 36% and most of the neutralizing antibody positive cases could neutralize HPV-6, and the titer of three samples was up to 2 560, which was consistent with the earlier finding that HPV-6 is the leading cause of genital warts [11].
The detection rate of HPV DNA was relatively high (60%), while the positive rates of HPV DNA have been reported to range from 16% to 20% in normal populations aged between 17 years to 59 years, while in patients with cervical cancer, the rate ranges from 83% to 96% [1, 13, 15, 17, 20, 21]. Therefore, the HPV infection rate in patients with genital warts in our study lay between the two different populations, with a detection rate of 54% for the high-risk types and 12% for the low-risk types. This was consistent with previous findings with detection rates of about 15% for the high-risk types and 3% for the low-risk types in normal populations and 84% for the high-risk types and 8% for the low-risk types in patients with cervical cancer. Therefore, the detection rates both for the high-risk and the low-risk types in genital warts patients were higher than that in normal populations, while the detection rate of the high-risk type in patients with genital warts was lower and the detection rate of the low-risk type was higher than that in the cervical cancer patients.
In our study, the most common types detected were HPV-68 (18%), HPV-16 (14%), HPV-58 (12%), HPV-33 (8%), HPV-6, HPV-11, HPV-18 and HPV-52 (6% for each). However, the types detected in the women's cervix do not necessarily indicate these types cause the warts, and we acknowledge that HPV6 is the leading cause of genital warts. The high-risk types, which were the most prevalent types detected here, may just be the most common cause of cervical infections, but may also indicate that the population has a higher risk of progression to cervical cancer.
The prevalence of HPV DNA types detected from the genital warts in our study differed to previous reports[11, 23], in which the detection rates were highest for HPV-6 and HPV-11. In contrast, the high-risk genotypes, such as HPV-68, HPV-16 and HPV-58 were more commonly detected than HPV-6 and -11 in our results. The reasons for the different HPV prevalence may be a result of differences in sample collection geographical regions or differences in methods used to obtain the cervical swab, which was used to detect and type HPV DNA. If the factors above can be excluded, we can speculate that HPV-6 neutralizing antibodies at a relative low level may be able to neutralize HPV-6, which may attenuate the detection rate of HPV-6 in swab samples. However, the HPV-6 neutralizing titers of the HPV-6 DNA negative cases was lower than that of the HPV-6 DNA positive cases, which is contradictory. Therefore, the function of HPV neutralizing antibodies induced by HPV natural infection needs further research.
Serum antibodies against HPV capsids may serve as markers for ongoing or previous infections[7, 14]. Regarding HPV-6, for instance, 26% (13/50) of cases positive for HPV-6 DNA or neutralizing antibodies were considered to be related to infection with HPV-6. Among these cases, 76.9% (10/13) were negative for HPV DNA and positive for neutralizing antibodies, suggesting that the majority of these infections had induced the production of sufficient neutralizing antibodies to clear the HPV infection, and the patients had recovered from the infection. However, 15.4% (2/13) of cases were positive for both HPV DNA and neutralizing antibodies, which indicates that either the infection was ongoing or the neutralizing antibodies had not cleared the viral infection. Another 7.7% (1/13) of cases were negative for neutralizing antibodies and positive for HPV DNA, which means that these infections cannot be detected by the presence of neutralizing antibodies. This may arise because the antibody titers decline over time among individuals with HPV infection. Another possible explanation for women failing to seroconvert is that they are genetically or immunologically predisposed not to recognize or respond to HPV infection [8].
Recently, PCR techniques have allowed the amplification of significantly longer DNA target sequences. We amplified five types of 8 kb genomes from the DNA samples extracted from the cervical swabs, which confirms that the PCR method used here can amplify various HPV genomes. With regards to the other types that could not be amplified, this may be a result of the poor quality of the DNA samples or the designed primers. The homologies were aligned with the whole genome sequence of the cloned PCR products. A PV type is defined as a complete viral genome with a sequence organization typical of PVs and an L1 gene sequence that is at least 10% dissimilar from that of any other PV type[12]. Each HPV type can be re-isolated in the form of genomic variants, which show a sequence variability of less than 2%[9, 22]. Between types and variants, subtypes were introduced and defined by a nucleotide sequence variability of 2% to 10% in the L1 ORF[6]. The homologies were aligned with the whole genome sequence of the cloned PCR products. In our study, alignment results showed that the sequence diversities were only about 1%-2%, which indicated the types of the genome we obtained (HPV-42, -52, -53, -58, -68) were a variant from their published isolate.














 DownLoad:
DownLoad: