-
Maize rough dwarf (MRD) is a devastating disease in northern China. The typical symptoms of MRD include stunting of plants, darkening of leaves, and white waxy swelling along the veins on the sheath and underside of the leaf blade. Infested plants usually produce poor or no head. In 2008, more than 700 000 hectares of maize (about one-fourth of the total planting area) in Shandong province was infected with incidence of 30%-50%. Some fields had an incidence of higher than 80%.
The virus causing MRD in China has been reported to be Rice black-streaked dwarf virus (RBSDV; genus Fijivirus, family Reoviridae)[3, 6, 26]. RBSDV infects rice, maize, wheat and other cereal hosts, and can be persistently transmitted by planthoppers including Laodelphax striatellus, Unkanodes sapporona and Unkanodes albifascia.
Southern rice black-streaked dwarf virus (SRBSDV), also known as rice black-streaked dwarf virus 2 (RBSDV2), is a novel member of the fijivirus group 2[15, 22, 27, 28]. The genome of SRBSDV contains ten segments of double-stranded RNA (dsRNA) named S1 to S10 with size of molecular weight in decreasing order [28]. SRBSDV was first observed to infect rice in Guangdong province in 2001, but now has become prevalent and caused serious rice losses throughout southern and central China, and northern Vietnam [22]. The major vector of SRBSDV is white-backed planthopper (Sogatella furcifera). Besides rice, SRBSDV also infects maize (Zea mays), barnyard grass (Echinochloa crusgalli), flaccid grass (Pennisetum flaccidum) and Juncellus serotinus [28]. In 2008, we detected SRBSDV from naturally infected maize plants in Ji'ning, Shandong province during an investigation of MRD. Here we report the nucleotide sequences of S7 to S10 of JNi4, a SRBSDV isolate from maize in northern China.
HTML
-
The isolate, Jni4, was isolated from a maize plant showing typical rough dwarf symptoms in a field in Ji'ning, Shandong province in 2008. Total RNA was extracted from infected maize leaves using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The cDNA was synthesized using primers listed in Table 1 and M-MLV reverse transcriptase (TransGen, Beijing, China) as described previously [3]. PCR amplification was conducted using LA Taq polymerase (TaKaRa Bio, Dalian, China) in 25 μL reaction systems, which contained 3 μL first-strand cDNA, 1.8 U TaKaRa LA Taq polymerase, 0.4 μmol/L of each primer, 2.5 μL 10× PCR buffer (containing Mg2+), and 200 μmol/L of each dNTP. The PCR amplification programs were preheating for 3 min at 94℃, 5 cycles of 1 min at 94℃, 50 s at 37℃, 2 min at 72℃, 30 cycles of 1 min at 94℃, 50 s at 56℃ (S7 and S9)/60℃ (S8)/51℃ (S10), 2 min at 72℃. The reaction was terminated with an extension of 72℃ for 10 min. Gene-specific primer pairs (GSPs) GSP7-5/GSP7-3 and GSP9-5/GSP9-3 were designed to amplify the 5′-and 3′-terminal regions of S7 and S9 with SMARTTM RACE cDNA Amplification Kit (Clontech, Mountain View, California, USA) following the manufacturer's instructions. Because the bands for S7 were weak or smeared in the agarose gel electrophoresis, the 5′-and 3′-terminal fragments of S7 were finally amplified by nested PCR with nested GSPs NGSP7-5 and NGSP7-3. The PCR products were purified using a DNA purification kit (TaKaRa, Dalian, China) and then cloned into pMD18-T vector (TaKaRa, Dalian, China).

Table 1. Primers used in this study
-
The positive clones with foreign inserts were screened with electrophoresis and PCR. For each segment, three or four clones from two separate PCR reactions were sequenced. In case of inconsistency, more clones were sequenced to obtain the consensus sequences[24]. Sequences were aligned using ClustalW and adjusted manually to confirm correct reading frame [11, 21]. Identities at nucleotide (nt) and amino acid (aa) levels were calculated with the MegAlign program of software DNAStar (DNASTAR, Inc., Madison, Wisconsin, USA). Phylogenetic trees were constructed and reliability was estimated with 1000 bootstraps using the neighbor-joining (NJ), minimum evolution (ME), maximum parsimony (MP), and unweighted pair-group method using arithmetic averages (UPGMA) using the MEGA4 software package (The Biodesign Institute, Tempe, Arizona, USA) [10, 20]. The viruses included for analysis are Fiji disease virus (FDV) [8, 14, 19], Oat sterile dwarf virus (OSDV) [9], Nilaparvata lugens reovirus (NLRV) [16-18], Mal de Río Cuarto virus (MRCV) [4, 5, 7], Maize rough dwarf virus (MRDV) [12, 13], Rice black-streaked dwarf virus (RBSDV) [23, 25] and two isolates of SRBSDV[22].
Virus isolate, RNA extraction and PCR amplification
Sequence determination, comparison and phylogenetic analysis
-
The S7 and S8 of JNi4 were 2176 bp (Accession No. HM998850) and 1928 bp (Accession No. HM 998851), respectively, and identical in length to those of SRBSDV isolates HN and GD. The S9 and S10 of JNi4 were 1899 bp (Accession No. HM998852) and 1797 bp (Accession No. GQ472845), respectively. Their untranslated region (UTR) was identical to GD in length but one nt less than HN in both the 5'-UTR of S9 and 3'-UTR of S10.
S7 contained two non-overlapping open reading frames (ORFs). ORFs 7-1 and 7-2 were 1074 bp and 930 bp, respectively, in length. S7 had a 5'-UTR of 40 bp, a 3'-NTR of 81 bp, and an intergenic region of 51 bp in length. S9 also contained two non-overlapping ORFs: ORF 9-1 1044 bp (52-1095) and ORF 9-2 630 bp (1159-1788). Both S8 and S10 contained one ORF only, which was 1776 bp (25-1800) and 1675 bp (22-1695) respectively. The termini of both S7 and S9 had conserved sequences of 5′-AAGTTTTT…… CAGCTGATGTC-3′. They also had a perfect or imperfect 7-9 bp inverted repeat immediately adjacent to the conserved terminal sequences, which is typical for a Fijivirus. The termini of S7 had a perfect inverted repeat of 5′-TTTCGACCT……AGGTCGA AA-3′, while the termini of S9 had imperfect inverted repeat of 5′-TAAGCCTGG……GCCGGCTTA-3′, which were consistent with previous reports [22, 27, 28] and which may act as an assembly signal during virus assembly [1]. Segments S8 and S10 also had terminal conserved sequences and inverted repeats, but those nucleotides were included in the primers and therefore not considered here.
-
The six ORFs of JNi4 and other fijiviruses shared identities of 18.2-85.0%, 13.0-61.6%, 16.1-71.9%, 14.9-78.1%, 14.7-73.2% and 18.1-84.7% at the aa level (Table 2); the highest identities with MRDV was for ORFs 7-1, 7-2 and 9-1 and with RBSDV for ORFs 8 and 10. JNi4 ORF 9-2 shared the highest nt identity with RBSDV, but highest aa identity with MRDV.

Table 2. The nucleotide/amino acid identities of JNi4 and other Fijiviruses between ORFs in S7 to S10
JNi4 shared identities of 99.7%, 99.1%, 98.9% and 98.6% with HN in the full-length genomic sequences of S7, S8, S9 and S10. The six ORFs (ORF7-1, ORF7-2, ORF8, ORF9-1, ORF9-2 and ORF10) of JNi4 shared aa identities of 99.7%, 99.7%, 99.2%, 97.7%, 99.0% and 99.1%, respectively, with those of HN. The S7 to S10 of JNi4 shared identities of 99.7%, 99.7%, 99.5% and 99.2%, respectively, with those of GD at nt levels; their six ORFs had aa identities of 99.7%, 100%, 99.8%, 99.4%, 100% and 99.6%, respectively.
-
JNi4 formed a separate clade with isolates GD and HN of SRBSDV in the phylogenetic trees constructed with S7 to S10 sequences (Fig. 1), indicating that JNi4 was an isolate of SRBSDV. These clades were were clustered to those of other Fijiviruses within the tree, confirming that SRBSDV was a distinct species in the genus Fijivirus. The phylogenetic trees constructed with the nucleotides and amino acids of six ORFs were consistent with those of full length genomic sequences.
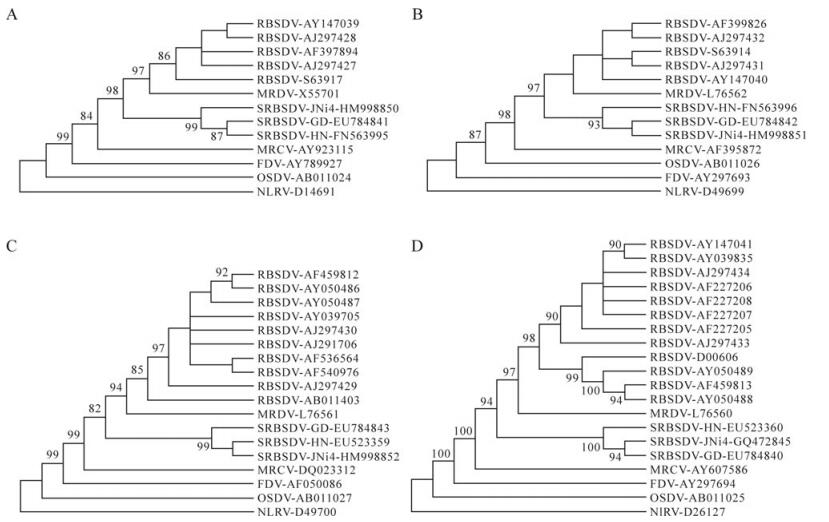
Figure 1. Neighbor-joining tree of SRBSDV isolate JNi and other fijiviruses constructed with genomic sequences of S7 (A), S8 (B), S9 (C) and S10 (D). The phylogenetic tree was also confirmed with methods including minimum evolution, maximum parsimony and unweighted pair-group methods using arithmetic averages available in the MEGA4.0 package. The phylogenetic tree was subjected to 1000 bootstrap tests. Branches with bootstrap values lower than 50% were collapsed. Only bootstrap values > 80% were shown. Isolates are indicated in the tree by virus name-accession number. The names of SRBSDV isolates were also listed.
Genome structure of S7 to S10
Nucleotide and amino acid identities
Phylogenetic analysis
-
Since its first observation in Guangdong in 2001[28], SRBSDV has become widespread in southern and central China and northern Viet Nam, and caused severe losses in rice production [22]. We investigated 106 maize samples from Shandong in 2008, and only detected SRBSDV from one sample, indicating that RBSDV is still the main cause of MRD in Shandong province. Because SRBSDV is primarily transmitted by white-backed planthopper (Sogatella furcifera) [28], which migrates routinely between north and south in many Asian countries including China[2], and JNi4 shared identities of > 98.6% with HN and GD in the genome of S7 to S10, we hypothesized that JNi4 was transmitted to maize plants in Ji'ning, Shandong occasionally by white-backed planthopper during their migration. During our detection of SRBSDV from maize in Shandong and neighboring provinces in northern China in 2009 and 2010, SRBSDV was found in samples from the same location (data not shown). SRBSDV was also detected from rice plants in Kumamoto Prefecture, Kyushu, Japan (information from Internet). However, this is the first report of SRBSDV from naturally infected maize in Shandong province, and is the northernmost region for the occurrence of SRBSDV in China to date. Considering the prevalence of SRBSDV in Northern Viet Nam and Southern China, we are paying close attention to the possible spread of SRBSDV and investigating its potential field hosts in northern China.














 DownLoad:
DownLoad: