-
As one of the most ubiquitous pathogens that has a worldwide distribution, herpes simplex virus type 1 (HSV-1) infects the respiratory or oropharyngeal mucosa and causes a variety of diseases ranging from mild illness to severe life-threatening infections such as herpes keratoconjunctivitis and herpes encephalitis. The primary infection in the mucosal epithelial cells is lytic while the virus gains access to the termini of sensory neurons and establishes life-long latent infections, which can transmit to the peripheral sites after reactivation and lead to recurrent lesions in the vicinity of the primary infected area. During productive infection, the linear, double stranded HSV-1 genome of approximately 152 kb encodes at least 84 proteins which are separated into three subsets known as immediate-early (IE), early (E), and late (L) proteins according to the sequential expression of the corresponding genes. More than half of these genes are poorly understood functionally [12, 13, 24] and identified as dispensable proteins for viral replication at least in some cell lines [1, 2]. These dispensable gene products are, however, demonstrated to be required for virus growth and pathogenesis in experimental animal systems [18, 20, 25, 31].
One such nonessential protein, UL4, has been identified and classified as a true late (γ2) gene product [7], but no functions have yet been assigned. Homologues of the UL4 gene are present in other members of the Alphaherpesviruses, such as the HSV-2 gene UL4 [23], the varicella-zoster virus gene 56 [3], the equine herpesvirus-1 gene 58 [33], the pseudorabies virus (PrV) gene UL4 [5] and the bovine herpesvirus-1 gene (BHV-1) UL4 [35]. Conservation of the gene suggests that it may play a certain role in the viral life cycle. Recent investigation has revealed that the PrV UL4 protein is a non-structural protein which is important for virus yields, release of mature virions, and enhancement of virion formation but it is not essential for PrV replication in vitro or in vivo [8].
ICP22 is an IE protein that enhances the expression of a subset of γ2 proteins. The UL4 protein has been identified as a putative nuclear protein for exhibiting co-localization with ICP22 in small, dense nuclear bodies in infection [14], suggesting the intriguing possibility of partial function overlap of ICP22 and the UL4 protein, and the UL4 protein may play certain roles related with ICP22 in the context of the small, dense nuclear compartment associated with late gene transcription. Furthermore, the late protein UL3 and UL20.5 are subsequently identified as the components of the discrete, dense nuclear spots [21, 36]. Since deletion of ICP22 resulted in a decreased accumulation of UL3 and, to a lesser extent, of UL4, and as neither UL3 nor UL4 localizes to the dense nuclear structures, with additional evidence showing that ICP22 by itself is sufficient to form small, dense nuclear bodies in transfection assay without other viral proteins, it has been suggested that co-localization of UL3 and UL4 is directed by ICP22 rather than by the interaction of the UL3 and UL4 proteins [21]. However, our recent systematic study showed determinately that both UL3 and UL20.5 are targeted to small, dense nuclear bodies by direct interaction with ICP22, whereas UL4 co-localizes with ICP22 through its direct interaction with UL3 but neither UL20.5 nor ICP22, and there is no interaction between UL3 and UL20.5 [40]. We also observed that UL4 is not a genuine nuclear protein as it localizes to sub-cytoplasmic structures when expressed alone. In addition, UL4 re-localized UL3 to the cytoplasm structure when co-expressed, while the subcellular distribution of UL4 was not altered when co-expressed with ICP22, but UL3 was redistributed in the nuclear foci identical to the subcellular localization pattern of ICP22, indicating that UL3 is the intermediate to bring UL4 to ICP22 and is required for the localization of UL4 in the nuclear compartment. Interestingly, the interaction between UL4 and UL3 took place in the cytoplasm where UL4 was located when expressed alone, prompting further investigation of the subcellular localization of UL4, which would be helpful to dig deeper not only into the interaction between UL3 and UL4, but also the role and mechanism of that interaction complex during the proliferation of HSV-1.
Active export of a protein from the nucleus depends on the presence of a specific nuclear export signal (NES) [10, 16]. To date three types of NES have been identified, for instance the ~10 amino acid leucine-rich NES initially identified in protein kinase inhibitor (PKI) and HIV-1 Rev, the M9 sequence of hnRNP A1, and a 24 amino acid stretch found in hnRNPK[28]. In addition, an increasing number of viral proteins, including HIV-1 Rev [16, 26, 27], HSV-1 UL47 [38], the RSV matrix protein [9] and BHV-1 protein BICP27 [6] have been found to contain a leucine-rich NES sequence, enabling the protein to be exported out of the nucleus. Chromosomal region maintenance 1 (CRM1; exportin 1), identified as an export receptor that recognizes NES directly, is responsible for the export of NES-containing proteins through the nuclear pore complex (NPC) using RanGTP [34, 37]. The pharmacological compound leptomycin B (LMB) directly interacts with CRM1 and blocks NES-mediated protein export.
In this study, we used living cell fluorescence microscopy, which has been extensively applied [4] and developed in our laboratory [11, 39, 41] to examine the distribution of heterologously expressed fusion proteins involving various deletions and point mutations of the HSV UL4 protein combined with enhanced yellow fluorescent proteins (EYFP) to identify the molecular determinants for subcellular localization of UL4; the molecular mechanisms underlying the nuclear export of UL4 was also elucidated.
HTML
-
COS-7 cells were grown in Dulbecco's modified MEM (DMEM; Gibco-BRL) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL).
-
All restriction enzymes used for cloning procedures were purchased from Takara (Dalian, China). T4 DNA ligase was from New England Biolabs (MA, USA). The HSV-1 (F) UL4 open reading frame was amplified with HSV-BAC DNA (pYEbac102) [32]. The primers for constructing all the plasmids are available upon request. All fragments were cloned into eukaryotic expression vector pEYFP-N1 (Clontech, BD Biosciences). Each construct was confirmed by sequencing. pRev-NES-EGFP was a generous gift from Dr. Gillian Elliott [38] and was used as the positive control for LMB treatment. pGEX-6p-1 Q69L Ran was a gift from Dr. Yoshinari Yasuda and RanQ69L dominant negative mutant was subcloned into pECFP-N1 (Clontech) to yield RanQ69L-ECFP.
-
Transfection and fluorescence microscopy experiments were performed as previously described [11, 39]. In brief, COS-7 cells were plated onto six-well plates (Corning, USA) in DMEM with 10% FBS at a density of 2.5×105 cells per well overnight before transfection. Transfection mixtures, consisting of 1.0 to 1.5 μg of plasmid and Lipofectamine Plus reagent (Gibco-BRL), were prepared according to the manufacturer's instructions. COS-7 cells were washed with Opti-MEM, and the transfection mixture, made up to 500 μL with Opti-MEM, was added to the cells and incubated at 37℃ in a humidified 5% CO2 incubator for 5 h. After that it was removed and replaced by an equal volume of DMEM containing 10% FBS. Cells were washed with fresh growth medium and processed for fluorescence microscopy 24 h posttransfection. When indicated, LMB (Sigma) was added to the culture medium at a final concentration of 10 ng/mL for 4 h prior to microscopy. The living cells were analyzed using fluorescence microscopy (Zeiss, Germany) 24 h after transfection, All the photomicrographs were taken under a magnification of 40×. Each photomicrograph shows a representative population of the cells with similar subcellular localization. Fluorescent images of EYFP and ECFP fusion proteins were presented in green and red pseudocolors, respectively. Images were processed with Adobe Photoshop.
Cells
Plasmids construction
Transfection, fluorescence microscopy and LMB treatment
-
The open reading frame of the HSV-1 (F) UL4 gene was located at the 5' end of the unique long (UL) region of the HSV-1 genome (Fig. 1a). The UL4 protein of HSV-1 (F) contained 199 amino acids corresponding to a molecular mass of 25 kDa [7]. The amino acid sequence of the UL4 protein was analyzed and predicted to have a hydrophobic, leucine-rich NES in the C-terminus, an N-terminal cleavage site for the mitochondrial presequence, and a KKXX-like motif 195KLLG198 in the C-terminus which may function as an ER retention signal (Fig. 1b).
-
Since fixation in an immunofluorescence assay may alter the localization of proteins, resulting in misleading conclusions in the analysis of the intracellular distribution of a specific protein, the living cells fluorescence microscopy technique [11, 39, 41] was applied in our study. To determine the exact subcellular localization of UL4 when expressed alone, the ORF of UL4 was inserted into pEYFP-N1 to generate UL4-EYFP (Fig. 2a) and the localization of the UL4-EYFP protein was imaged in transfected living cells (Fig. 2b). As shown in Fig. 2b, UL4 is exclusively restricted to cytoplasmic granulo-like structures, suggesting that there might be an NES, given that UL4-EYFP is small enough ( < 60 kDa) to freely diffuse through the aqueous channel delimitated by the NPC embedded in the nuclear membrane. According to the analysis of amino acid sequence, there is a cluster of leucine-rich residues between amino acids 178 and 186, namely 178VEVLREIQL186, resembling the most prevalent NES in which the leucine residues are critical for function [10, 22].
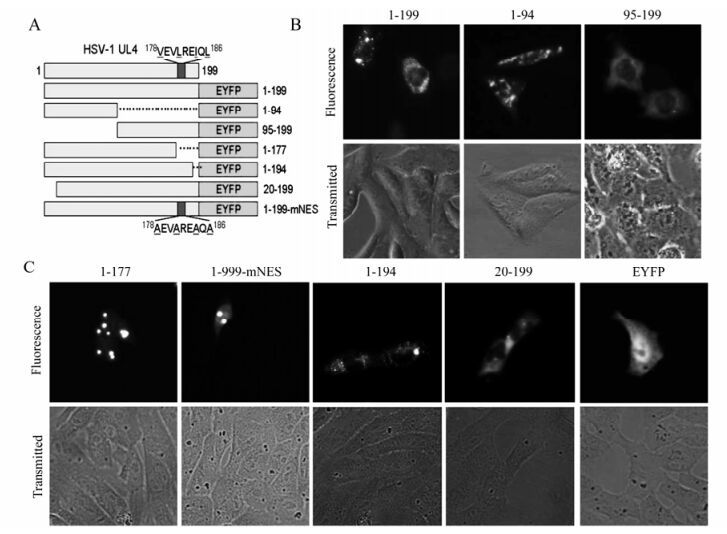
Figure 2. Mapping of the subcellular localization signal in the UL4 protein. A: Schematic diagram of wild-type UL4 and the mutants fused with EYFP. B: Subcellular localization of UL4(1-199)-EYFP and deletion mutants aa1-94-EYFP and aa95-199-EYFP. Each image is representative of the vast majority of the cells observed. C: Subcellular localization of the other UL4 mutants fused with EYFP. Representative fluorescence images of the vast majority of living cells for indicated EYFP fusion proteins and EYFP fluorescence were analyzed in living cells 24 h after transfection. Light-translucent pictures showed cellular morphology.
To map the amino acid sequence responsible for the localization of UL4, a series of deletion mutants encompassing different segments of UL4 fused with EYFP were constructed (Fig. 2a). As shown in Fig. 2b, mutant aa95-199-EYFP accumulated mainly in the cytoplasm whereas mutant aa1-94-EYFP showed a different localization pattern. Furthermore, mutant aa1-177-EYFP demonstrated diffuse fluorescence throughout the nucleus and cytoplasm but with several bright aggregation spots (Fig. 2c), suggesting that an NES exists in the C-terminus of UL4 as predicted. To further validate the leucine-rich 178 VEVLREIQL 186 actually acted as a functional NES for the export of UL4 out of the nucleus, the putative NES was mutated by replacing the leucine and hydrophobic amino acid residues with neutral alanine residues to create 178AEVAREAQA186 (1-199-mNES -EYFP; Fig. 2c). Fluorescence results for aa1-199-mNES-EYFP showed identical localization with the pattern observed for aa1-177-EYFP. Taken together, these results indicated that there was an NES spanning aa 178 to 186 in the C-terminus of UL4 and within which the leucine and hydrophobic amino acid residues played a key role.
Interestingly, the UL4-EYFP did not show an even distribution in the cytoplasm, but enriched in cytoplasmic granulo-like structures, and aa1-177-EYFP and aa1-199-mNES-EYFP also contained several bright aggregation spots, implying that there might be additional subcellular organelle location signals. The examination of the UL4 amino acid sequence suggested that two signals relevant to the mitochondrion and ER respectively might be the cause of the granulo-like location pattern. To identify the molecular determinants, deletion mutants aa1-194-EYFP and aa20-199-EYFP were constructed and the fluorescence in transfected COS-7 cells were examined. As shown in Fig. 2c, mutant aa1-194-EYFP with the deletion of putative ER membrane retention signal had the same distribution pattern as UL4-EYFP, indicating the predicted ER targeting signal 195KLLG198 can not act as an ER membrane retention signal and target the UL4 to ER membrane. Conversely, deletion of the 19aa at the N-terminus leads to the loss of the cytoplasmic granulo-like pattern and these mutants targeted the cytoplasm in a diffuse pattern. We concluded from these results that the N-terminal 19 amino acid residues were required for the granulo-like cytoplasmic pattern of UL4.
-
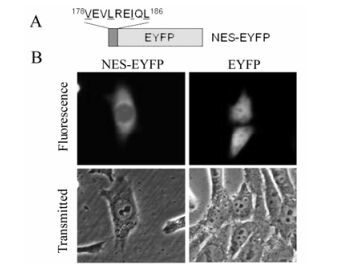
To determine if the putative UL4 leucine-rich NES is sufficiently functional and would be transferable to mediate nuclear export of heterologous proteins in the same manner as HIV-1 Rev NES. A DNA fragment corresponding to the amino acid sequence 178VEVLREIQL186 which represents the putative NES, was synthesized and ligated to the N-terminus of the EYFP DNA sequence, yielding NES-EYFP (Fig. 3a). As expected, NES-EYFP was targeted more to the cytoplasm than the nucleus (Fig. 3b), suggesting that to some extent, the leucine-rich sequence is a functional NES.
-
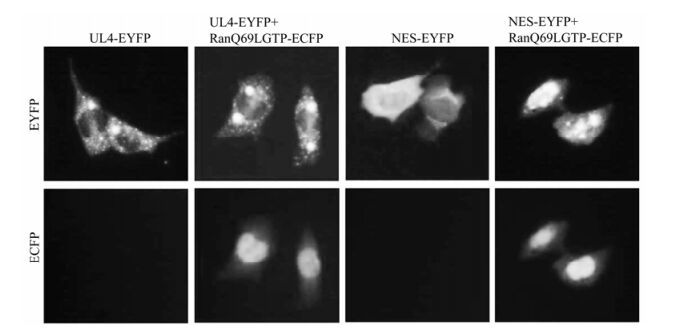
Leucine-rich NES has been identified in an increa sing number of cellular and viral proteins with quite heterologous biological functions. As the best characterized exportin, CRM1 mediates nuclear export of many classical NES-containing proteins as well as certain mRNAs and spliceosomal U small nuclear (sn) RNAs. Its activity can be specifically inhibited by the drug LMB [17, 29], a cytotoxin that is shown to bind to CRM1. Binding of RanGTP to the NES-containing protein-CRM1 complex is indispen sable and is involved in the hydrolysis of GTP into GDP during nuclear export. To elucidate the nuclear export mechanism of UL4, LMB treatment was performed after transfection of pUL4-EYFP, pNES-EYFP and positive control pRev-NES-EGFP. The results showed that the nuclear export activities of UL4 were affected by treatment with LMB at 10 ng/mL for 4 h, completely abrogating its exclusively cytoplasmic granule-like localization and the majority of cells exhibited a distribution throughout the nucleus and the cytoplasm in a totally different pattern (Fig. 4), ndicating the export of the UL4 is CRM1 dependent. Additionally, COS-7 cells are co-transfected with the dominant negative Ran protein, RanQ69LGTP, which is deficient in GTP hydrolysis, and UL4-EYFP or NES-EYFP, respectively. Subcellular localization of these mutants of UL4 and RanQ69LGTP was observed. As shown in Fig. 5, cytoplastic accumu-lation of UL4-EYFP and NES-EYFP were altered when co-transfected with RanQ69LGTP, which suggests the Ran protein and GTP hydrolysis are involved in CRM1-meadiated nuclear export of UL4. This evidence demonstrates that UL4 is exported from the nucleus to the cytoplasm in the CRM1-dependent pathway involving RanGTP hydrolysis.
Molecular characterization of the UL4 protein
Mapping of the subcellular localization signals in the UL4 protein
The identified leucine-rich sequence is the functionally transferable NES of UL4
Molecular mechanism for the export of UL4
-
As a real late HSV-1 protein with no assigned function, UL4 has been identified as a nonessential protein which is not necessary for viral replication in cell culture [1]. The conservation of the UL4 gene in Alphaherpesviruses, however, suggests that UL4 might play a more relevant role in vivo. It was reported later that this protein is dispensable for latency, reactivation and pathogenesis in mice [15] and a study was carried out on recombinant HSV-1 with mutated UL4 (UL4 mutant HSV-1) that retained significant portions of the ORF that might still allow the expression of truncated UL4 protein containing the N-terminal 60 amino acids. The UL4 mutant HSV-1 exhibited only minor replication defects in vitro, and pathogenesis as well as latent infections in mice were also not significantly affected [15]. However, it could not be excluded that the truncated UL4 was expressed and partly functional in the UL4 mutant HSV-1 and that deletion of the entire gene would lead to a more pronounced phenotype, it's likely that wild type UL4 has additional contributions during viral life cycle, as is the fact that it targeted to the small, dense nuclear compartment related to late gene transcription with ICP22, a multifunctional viral regulator, UL3 and UL20.5 during infection.
In this study, for the first time we characterized the subcellular localization of HSV-1 UL4 without other viral protein, which was prompted by the previously described novel interaction between UL4 and UL3 in the cytoplasm where UL4 is located. In the present study, we have shown that the UL4 protein of HSV-1 is exported out of the nucleus, but the exact relevance of this to the interaction with UL3 or its localization in the nuclear bodies during infection is yet to be demonstrated. The putative leucine-NES in the C-terminus directs the export of UL4 from the nucleus and constrains it within the cytoplasm since deletion mutant aa1-177-EYFP with deletion of C-terminus was detected in the nucleus, which is similar to the localization of 1-199(mNES)-EYFP with hydrophobic residues replaced by neutral alanine residues. The difference of the location pattern between these two mutants with EYFP (Fig. 2c) can be attributed to the N-terminal signal in UL4. This N-terminal signal is a cleavable presequence which targets the mitochondrial matrix, inner membrane, and intermembrane space, and was identified by the observation that when this presequence was deleted, the mutant abandoned the granulo-like cytoplasmic localization pattern and was distributed diffusely throughout the cytoplasm. However, the exact distribution of UL4 in the mitochondrion is still to be determined.
Given that the leucine-rich NES 178VEVLREIQL186 mediates the export of wild type UL4 out of the nucleus, the NES would be able to direct the nuclear export of heterologous proteins, eg. EYFP. However, as shown in Fig. 3b, NES only slightly targeted EYFP to the cytoplasm. This may be because NES contains only a few amino acid residues and could be folded inside of the fusion protein NES-EYFP, consequently blocking binding of the NES to exportin [30].
In addition, we demonstrated that the nuclear export activity of the functional NES was completely blocked by the specific nuclear export inhibitor LMB, indicating that the NES mediated the nuclear export of UL4 via the export receptor CRM1. Finally, we also demonstrated that Ran protein and GTP hydrolysis are involved in the CRM1-meadiated nuclear export.
















 DownLoad:
DownLoad: