-
Influenza virus surface glycoprotein neuraminidase (NA) is the primary target of influenza for structurebased drug design (von Itzstein M, et al., 1993). To date, the most effective anti-influenza NA drugs (NAIs) include oseltamivir (Tamiflu) and zanamivir (Relenza) that are specific for influenza A and B viruses (Gubareva L V, et al., 1995; Hata K, et al., 2008; Ryan D M, et al., 1995; Woods J M, et al., 1993). However, virus variants associated with drug-resistance have arisen from treatment with current NAIs, which include drugresistant seasonal H1N1 and H3N2 human influenza viruses as well as drug-resistant highly pathogenic avian influenza (HPAI) viruses of the H5N1 subtype. Emergence of drug-resistant influenza viruses undermines our current drug therapies.
Oseltamivir treatment has led to influenza A virus drug-resistant variants within N1, N2 and N9 subtypes as well as influenza B virus variants. Clinical human influenza A/H3N2 and A/H1N1 isolates recovered from oseltamivir-treated individuals contain R292K, E119V and N294S substitutions in NA N2 and H274Y and N294S substitutions in NA N1. Furthermore, studies of drug-treated clinical H5N1 isolates and recombinant H5N1 viruses have demonstrated reduced sensitivity to oseltamivir (de Jong M D, et al., 2005; Le Q M, et al., 2005; Yen H L, et al., 2007). In this report, we have adapted a cell-based assay to investigate NA susceptibility to oseltamivir. Several catalytic residue substitutions have been implicated in oseltamivir resistance and we have found that a handful of these residues display varying phenotypes in an NA N1 background shedding light into subtype specific effects on particular mutations of the enzyme active site associated with altered NAI susceptibility. Using a lentiviral-based pseudotyping system, we have demonstrated inhibition of HIV/HA pseudoviral particles from producer cells in the presence of oseltamivir permitting a resourceful screen to assess altered NAI susceptibility of various NA mutants. The utility of this system may provide a platform to screen novel therapeutics in influenza viral infection.
HTML
-
The HA gene used in this study was isolated from a highly pathogenic avian influenza virus in China, A/Goose/Qinghai/59/05 (H5N1) (Liu et al., 2005). NA cDNA from the mouse-adapted A/Puerto Rico/8/1934 (H1N1) virus was kindly provided by John C. Olsen (University of North Carolina, Chapel Hill). The HIV-1 proviral vector pNL4-3.Luc.R-E-was obtained through the NIH AIDS Research and Reference Reagent program (He et al., 1995) NA inhibitor, D-tartrate salt of (3R, 4R, 5S)-4-acetylamino -5-amino-3-(1-ethyl-propoxy)-cyclohex-1-enecarboxyl ic acid with 1.5 hydrate (oseltamivir carboxylate; RO0640802-002) was kindly provided by F. HoffmannLa Roche Ltd. (Basel, Switzerland). Oseltamivir carboxylate was shipped as a white, crystalline solid. The chemical was dissolved in sterile water to a concentration of 10 mg/mL, filter sterilized using a 0.22-μm-pore size filter (Nalgene) and then serially 10-fold diluted. Human 293T cells were maintained in DMEM (HyClone) supplemented with 10% FBS (HyClone), 100 μg/mL of streptomycin and 100 units of penicillin (Invitrogen).
-
Amino acid substitutions were generated by sitedirected mutagenesis using the Strategene QuickChange mutagenesis kit following the supplier's protocol. All mutations were confirmed by DNA sequencing of the full-length NA gene.
-
Influenza pseudovirions were produced by transient co-transfection of human 293T cells using a polyethylenimine (PEI)-based transfection protocol (Godbey W T, et al., 1999). Plasmids encoding hemagglutinin (HA), neuraminidase (NA) and replication-defective HIV vector (pNL4.3. Luc-R-E-or pNL4.3.GFP-R-E-) were used for transiently co-transfection on 293T producing cells. Seven hours after transfection, cells were washed by phosphatebuffered saline (PBS), 6 mL of fresh medium was added to each plate. Forty-eight hours post transfection, the supernatants were collected and filtered through 0.45 μm pore size filter (Nalgene) and were used directly for infection. The remaining pseudovirions were stored at -80 ℃ until further use.
Target 293T cells were seeded into 24-well plates at a density of 5 × 104 cells/well 24 h prior to infection. Five hundred microliters of pseudovirus-containing supernatant was added to the target cells and incubated overnight. A titration of oseltamivir carboxylate was added directly to the culture medium of 293T producer cells twice during pseudovirion production, both at 20 h and 44 h post-transfection. Susceptibility of influenza pseudovirus to oseltamivir carboxylate was assessed by generating influenza pseudovirus in 293T producer cells in the presence of increasing concentrations of oseltamivir ranging from 1.0 nmol/L to 1.0 μmol/L. Relative infectivity was determined 48 h post-infection by expression of the luciferase reporter in the infected cell lysates using a luciferase assay kit according to the supplier's protocol (Promega). Luciferase activity was measured using an FB12 luminometer at 10 second intervals (Berthold detection system). Each sample was done in triplicate and the experiments repeated at least three times.
-
Producer cell culture supernatants were harvested 48 h post-transfection, filter-sterilized and concentrated over a 30% Sucrose-NTE cushion consisting of 100 mmol/L NaCl, 10 mmol/L Tris (pH 7.4), and 1 mmol/L EDTA. The samples were spun at 55 000 rpm for 2 h in a SW55Ti rotor at 4℃. Pseudovirion pellets were resuspended in 50μL of Tris-buffer containing 50 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, and 5 mmol/L EDTA. Two fold serial dilutions were mixed with an equal volume of 0.5% chicken erythrocyte suspension (CRBCs; Lampire Biological Laboratories) and incubated in a U-bottomed 96-well plate at 4 ℃. HA titers were recorded after 1 h. Hemagglutination assay experiments were repeated at least three times.
-
A standard fluorometric enzyme assay developed by Potier et al. (Potier et al., 1979) was adapted to measure NA activity. Producer cell lysates transfected with representative NA constructs were added to NA fluorogenic substrate 2'-(4-methylumbelliferyl-Nacetylneuraminic acid (4-MUNANA; Sigma) to a final concentration of 100 μmol/L. The reactions were carried out in 50 μL of 33 mmol/L MES (pH 6.5) containing 4 mmol/L CaCl2 in 96-well black Optiplates (BD Biosciences). A titration of oseltamivir carboxylate (ranging from 0.5 nmol/L to 128 nmol/L) was added to the reaction mixtures and then incubated in a 37 ℃ water bath for 1 h. The reactions were terminated by adding 150 μL stop solution containing 0.5 mol/L NaOH (pH 10.7) and 25% ethanol. The fluorescence of released 4-methylumbelliferone was measured using a Labsystems Fluoroskan Ⅱ spectrophotometer (PerkinElmer). The excitation wavelength was set at 355 nm, and the emission wavelength was set at 460 nm. Samples were done in triplicate and the experiments repeated at least three times.
Constructs, reagents and cell lines
Site-directed mutagenesis of influenza neuraminidase gene
Production of pseudovirions and infection assay
Hemagglutination assay
Neuraminidase enzyme assay
-
Resistance to neuraminidase inhibitors (NAIs) continues to increase as the influenza virus acquires new mutations, and a quick screening system is crucial to assess possible pandemic threatening mutations that confer resistance to globally stockpiled antivirals. To meet this goal, we have employed a lentivirus-based pseudotyping system to study the effects of various substitutions in NA on oseltamivir sensitivity.
Lentiviral-based reporter viruses pseudotyped with an avian influenza hemagglutinin (HA) glycoprotein were generated in human 293T producer cells by transient co-transfection of an HA-expression plasmid along with an env-deficient HIV-1 vector (pNL4-3-Luc-R--E-) carrying a luciferase reporter gene. Susceptible target cells were challenged with the producer cell culture supernatants collected 48 h post-transfection and transduction was determined by luciferase activity in the target cells. Inclusion of an NA gene from a mouse-adapted human virus strain (PR8), heretofore known as NAH, resulted in efficient transduction, 1.7 × 107 RLU (Fig. 1A).
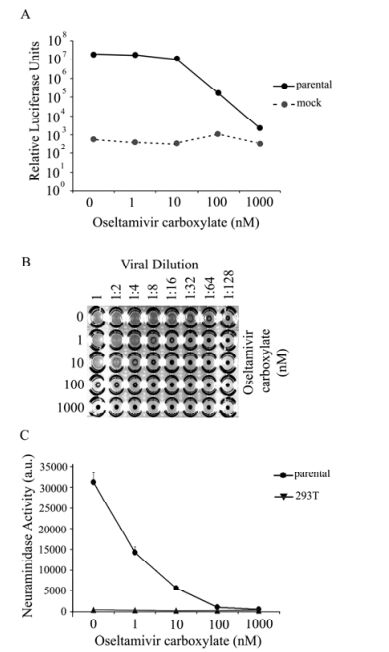
Figure 1. Inhibition of parental NA activity by oseltamivir. HIV/HA pseudovirions were produced by 293T cells in the absence or presence of oseltamivir and release was measured through ability of producer cell supernatants to (A) infect 293T target cells as measured via luciferase activity in the target cell lysates. Data shown is representative data from several experiments, each done in triplicate. Background luciferase levels were measured from lysates of target cells transduced with supernatant from producer cells lacking NA in their transfection. B: As another measure of pseudovirion release, a hemagglutination assay was done using equal volumes of chicken red blood cells and purified pseudovirus collected from producer cells. Virus was serial diluted out until 1:128. PBS was used as a negative control. C: NA enzymatic activity was directly measured in the absence or presence of oseltamivir by ability of producer cell lysate to cleave 2'-(4-methylumbelliferyl)-N-acetylneuarminic acid substrate and measured. Lysates of untransfected cells were used as a control. Error bars represent standard deviation. Data shown is representative data from several experiments, each done in triplicate.
HA/NAH-mediated luciferase levels exhibit a dosedependent decrease with increasing concentrations of oseltamivir carboxylate (OC) added to the culture medium of 293T producer cells during HIV/HA pseudovirion production (Fig. 1A). In the absence of inhibitor, resulting luciferase activity was more than 1 × 107 RLUs for 293T cells. A marginal decrease in luciferase levels was observed in the presence of 1 nmol/L OC. Luciferase levels started to decrease more dramatically in the presence of 10 nmol/L or higher OC compared to no drug. In the presence of 100 nmol/ LOC, luciferase activity dramatically decreased more than 100-fold, and the levels further decreased in the presence of 1 μmol/L OC comparable to background levels in this system (Fig. 1A). Thus it is clear that presence of OC in the producer 293T cells reduced the transduction ability of the HIV pseudovirions, consistent with the notion that OC inhibits viral particle release of HIV/HA pseudovirions.
To directly measure viral particle release, the HIV/HA pseudovirions in the presence or absence of OC were collected, concentrated and used in a hemagglutination assay. In the absence of OC, even at 1:64 viral dilution, hemagglutination activity was observed (Fig. 1B, top row). In contrast, in the presence of 1 000 nmol/L of OC, no hemagglutination activity was observed even without any viral dilution (Fig. 1B, bottom row). Furthermore, a reverse OC dose-dependent hemagglutination activity was observed (Fig. 1B, comparing rows with 1 nmol/L to 1 000 nmol/L). These results demonstrate that OC directly inhibited viral particle release of HIV/HA pseudovirions from the 293T cells.
The NA enzymatic activity and its inhibition by OC were assayed by the ability of NA to cleave a 2'-(4-methylumbelliferyl)-N-acetylneuraminic acid substrate (4-MUNANA). Cell lysates from the 293T producer cells untreated with OC were cleared and incubated with 4-MUNANA substrate and either with or without OC, and cleavage was measured fluorometrically. Without OC, the enzymatic activity of NA was approximately 30 000 arbitrary units (a.u.), and the level dropped nearly 50% with the 1nM of OC (Fig. 1C). At the higher concentrations (100 nmol/L or 1 000 nmol/L), the enzymatic activity of NA was completely abolished. Therefore, together with the aforementioned results, these results demonstrate that the HIV-based pseudotyping assay system can be used as a sensitive surrogate assay system for NA to oseltamivir.
-
The most commonly reported NA mutation that confers oseltamivir resistance in subtype N1 is H274 substitution, which is a structural residue in the active site (Fig. 2). To further validate the HIV-based pseudotyping system as a surrogate assay for NA resistance to OC, first we used site-directed mutagenesis to incorporate the H274Y substitution in the parental NA and performed the same assays as described above. Mutant H274Y mutant behaved like the parental NA at 0, 1, and 10nM concentrations of OC; in contrast, at 100 nmol/L and 1 000 nmol/L of OC, the HIV/HA pseudovirions with mutant H274Y gave at least 100-fold more transduction (measured by luciferase levels) compared with the parental NA (Fig. 3A), consistent with the notion that H274Y is resistant to OC.
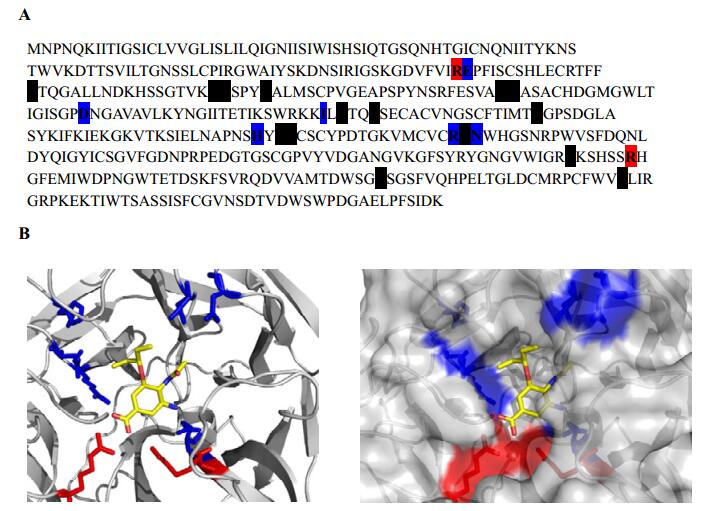
Figure 2. NAH catalytic residues within the active site. A: NA amino acid sequence. Targeted residues E119, I222, D198, H274, and N294 are colored blue. Additional catalytic residues are shaded black. B: Left panel, ribbon diagram of NA active site with mutation sites labeled blue and catalytic residues R118 and R371 colored red. Right panel, ribbon and surface representation of NA active site. Oseltamivir carboxylate is displayed in yellow.
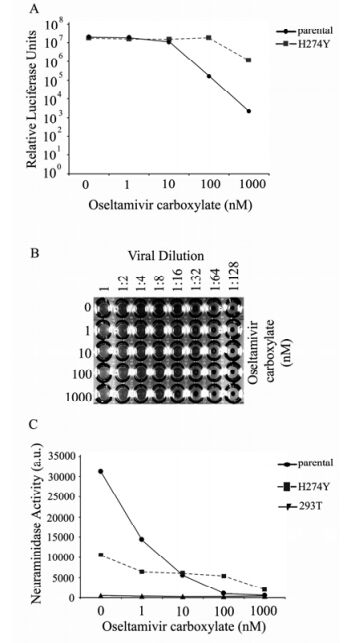
Figure 3. NA substitution H274Y confers resistance to oseltamivir carboxylate inhibition. A: Transduction of HIV/HA pseudovirions expressing NA H274Y. Relative infectivity is determined by luciferase activity (relative light units RLUs) from infected 293T target cell lysates. Values depict the average of triplicate samples. B: Hemagglutination activity of HIV/HA pseudovirions expressing NA H274Y Cell culture supernatants were harvested 48 h post-transfection. Mixtures of concentrated pseudovirions and chicken red blood cells were incubated at 4 ℃ and HA titers recorded 1 h later. PBS served as a negative control. C: Analysis of NA H274Y enzymatic activity measured by the release of 4-MUNANA.
Next, a hemagglutination assay was performed to evaluate how mutant H274Y behaved in the presence of OC (Fig. 3B). It is clear that mutant H274Y was very resistant to OC in its ability to release the HIV/HA virions in stark contrast to the parental NA (comparing Fig. 3B with Fig. 1B). For mutant H274Y, even at the OC concentration of 1, 000 nM, hemagglutination was observed when the viruses were diluted at 1:4 (the bottom row of Fig. 3B). These results demonstrate that mutant H274Y can function more efficiently than the parental NA in mediating HIV/HA particle release.
Interestingly, the enzymatic activity of mutant H274Y, as measured by substrate cleavage, was notably lower (about one third of the parental NA) than the parental NA in the absence of OC (Fig. 3C, and Table 1). Also it was observed that mutant H274Y displayed resistance to OC since its enzymatic activity did not decrease in a dose-dependent manner like the parental NA (Fig. 3C). These results are totally consistent with the behavior of mutant H274Y in response to oseltamivir reported by others.
NA Construct Oseltamivir
(nmol/L)Luciferase Activity
(RLU)HA
(U/mL)4-MUNANA Release
(a.u.)Parental 0 1.8(±0.01)×107 32 3.1(±0.2)×104 1 1.8(±0.06)×107 4 1.4(±0.1)×104 10 1.1(±0.2)×107 2 5.6(±0.4)×103 100 1.6(±0.09)×105 0 1.0(±0.04)×103 1000 2.2(±1.3)×103 0 4.9(±0.3)×102 H274Y 0 1.8(±0.09)×107 8 1.0(±0.1)×104 1 1.6(±0.05)×107 16 6.4(±0.4)×103 10 1.5(±0.1)×107 16 6.0(±0.4)×103 100 1.8(±0.02)×107 8 5.3(±0.1)×103 1000 1.1(±0.3)×106 4 1.9(±0.2)×103 N294S 0 1.8(±0.05)×107 4 1.1(±0.06)×104 1 1.8(±0.03)×107 4 1.2(±0.07)×104 10 1.8(±0.03)×107 4 1.1(±0.1)×104 100 4.0(±1.2)×106 4 5.7(±0.2)×103 1000 2.2(±0.7)×105 0 1.3(±0.06)×103 D198N 0 5.7(±2.1)×105 2 7.0(±0.5)×102 1 2.6(±0.4)×105 2 6.6(±0.2)×102 10 4.0(±1.3)×104 0 5.1(±0.2)×102 100 1.6(±0.2)×104 0 3.3(±0.2)×102 1000 1.1(±0.3)×104 0 2.5(±0.08)×102 I222L 0 1.5(±0.2)×107 8 5.1(±0.4)×103 1 1.5(±0.2)×107 8 4.4(±0.2)×103 10 1.1(±0.1)×107 2 3.6(±0.2)×103 100 1.4(±0.3)×105 0 1.1(±0.2)×103 1000 4.1(±1.5)×104 0 3.1(±0.08)×102 E119V 0 2.5(±1.8)×104 0 5.7(±0.7)×102 1 3.3(±2.9)×104 0 4.0(±0.2)×102 10 7.6(±4.3)×104 0 4.3(±0.1)×102 100 2.0(±2.6)×104 0 3.5(±0.3)×102 1000 7.4(±0.3)×103 0 2.7(±0.5)×102 R292K 0 4.9(±1.4)×103 0 6.4(±0.3)×102 1 1.1(±0.1)×104 0 4.1(±0.2)×102 10 8.1(±3.8)×103 0 4.3(±0.5)×102 100 4.7(±0.53)×103 0 4.1(±0.7)×102 1000 1.4(±0.32)×104 0 4.6(±0.1)×102 Table 1. Summary of Characteristics of NA mutants
Therefore, we conclude that the HIV-based pseudotyping assay is a reliable surrogate system to study oseltamivir resistance of influenza viruses.
-
In addition to mutant H274Y of NA, other NA mutants have previously been reported to show oseltamivir resistance (Fig. 2). N294S has been reported to show resistance in subtype N1; R292K, E119V, and I222L have been reported as showing resistance in subtype N2; D198N has been shown to demonstrate resistance in influenza B. These NA mutants were evaluated in their oseltamivir resistance profiles using the HIV-based pseudotyping assay described above.
R292K and E119V. These substitutions were among the earliest reported as showing oseltamivir resistance to subtype N2 NA. In this study, using a subtype N1 NA, the pseudovirion release as measured by luciferase was equivalent to the background level and the thus oseltamivir had no effect at any concentration (Fig. 4A). There was also no observable hemagglutination activity (Table 1). Furthermore, the neuraminidase activity as assayed in the cleavage assay was equivalent to the background level, and oseltamivir had no effect at any concentration (Fig. 4B). Taken together, these results suggest that these mutations render the subtype N1 NA enzymatically inactive.
Figure 4. NA substitutions displaying varying sensitivities to oseltamivir carboxylate inhibition. A: Transduction potential of HIV/HA pseudovirions expressing NA substitution mutants. Relative infectivity is determined by luciferase activity (relative light units RLUs) from infected 293T target cell lysates. Values depict the average of triplicate samples. Error bars indicate standard deviations. B: Analysis of NA substitution mutant enzymatic activity measured by the release of 4-MUNANA. C: Relative NA enzymatic activities of NA substitutions in response to different concentrations of oseltamivir carboxylate based on the data from (B).
N294S. When this NA mutant was evaluated by the transduction with the HIV/HA pseudovirions produced in the 293T producer cells, mutant N294S behaved like mutant H274Y (Fig. 4A), consistent with the reports that both mutants are resistant to OC in the N1 subtype. The hemagglutination assay showed lower HA titers of mutant N294S in the absence of OC, but these levels were maintained at 100 nmol/L of OC, contrasting to that of the parental NA which displayed no hemagglutination activity at such a OC concentration (Table 1). The enzymatic activity of mutant N294S mimicked that of mutant H274Y (Fig. 4B and 4C). Therefore we conclude that mutant N294S, like mutant H274Y, is resistant to oseltamivir.
D198N. This mutant was found to display an intermediate phenotype when it was evaluated in the HIV/HA pseuodtyping transduction assay without OC treatment, while it showed a dose-dependent sensitivity to OC treatment (Fig. 4A). Very weak or no hemagglutination activity was detected for this mutant (Table 1). The enzymatic activity of this mutant was really low (Fig. 4B and Table 1) compared to the parental NA. These results indicate that D198N substitution renders NA enzymatically inactive.
I222L. This mutant behaved like the parental NA in its OC profile except at 1 000 nmol/L, measured by transduction, suggesting that this mutant is somewhat resistant to OC treatment (Fig. 4A). The HA activity of this mutant was lower compared to the parental NA (Table 1), and the enzymatic activity of this mutant was approximately 1/6 that of the parental NA, and showed some resistance to OC treatment (Fig. 4B and 4C). These results indicate that this mutant is somewhat deficient in the enzymatic activity, and displays a slight oseltamivir-resistant phenotype.
HIV/HA pseudovirion release is inhibited in the presence of oseltamivir carboxylate
H274Y substitution of NA is resistant to oseltamivir carboxylate
Different NA mutants behave differently in their resistance to oseltamivir carboxylate
-
Reports that NAI resistance is on the rise in seasonal influenza viruses have prompted concerns over the current anti-influenza therapeutics. The WHO reported in 2008 that seasonal influenza H1N1 viruses are growing more resistant to oseltamivir (WHO, 2008). Treatment with oseltamivir carboxylate—a potent and selective inhibitor of influenza A and B viruses—can select for virus variants that are resistant to NAI inhibition. Molecular analysis of clinical isolates recovered from oseltamivir-treated individuals identified variants containing substitutions within the NA active site (Yen H L, et al., 2006). Human influenza H1N1 and H3N2 viruses and HPAI H5N1 variants containing H274Y and N294S mutations have been recovered from oseltamivirtreated individuals (de Jong M D, et al., 2005; Gubareva L V, et al., 2001; Kiso M, et al., 2004; Le Q M, et al., 2005; Weinstock D M, et al., 2003; Whitley R J, et al., 2001). Additionally, in vitro analysis of viral replication in the presence of oseltamivir as well as NAI treatment in infected-animal models have also demonstrated selection of virus variants with reduced susceptibility to inhibitors (Gubareva L V, et al., 2001; Gubareva L V, et al., 2001; Kiso M, et al., 2004; Le Q M, et al., 2005; Whitley R J, et al., 2001).
The most common substitution associated with oseltamivir-resistance of NA N1 and N2 subtypes are H274Y and R292K, respectively. Substitution of E119 is frequently observed with N2 resistance and a variety of amino acid substitutions are stably maintained including valine, alanine and aspartic acid at this position (Baz M, et al., 2006; Ison M G, et al., 2006; Kiso M, et al., 2004; Whitley R J, et al., 2001). Influenza B viruses have also demonstrated reduced susceptibility to oseltamivir treatment and variants have contained D198N and I222T substitutions within NA. Based on these reports, we characterized several NA substitutions implicated in oseltamivir resistance in the context of NA N1: E119V, D198N, I222L, H274Y, N294S and R292K. These NA substitutions displayed interesting properties and can be grouped into three classes according to: (Ⅰ) NAI-resistance (H274Y and N294S), (Ⅱ) NAI-sensitivity (D198N and I222L), and (Ⅲ) compromised NA function (R292K and E119V).
To evaluate whether these substitutions exhibit altered sensitivity to oseltamivir, we adapted an HIV-based cell assay and monitored inhibition of HIV/HA production in the presence of oseltamivir. Production of HIV/HA pseudovirions, measured by reporter virus agglutination of CRBCs and luciferase activity, is used for the readout of NA function. Thus, obstruction of pseudoviral particle release is evidenced by a decrease in luciferase levels and a corresponding loss of hemagglutination. The parental NA mediates efficient production of HIV/HA pseudovirions as evidenced by high levels of luciferase activity and hemagglutination. We have shown that oseltamivir carboxylate effectively blocks the release of HIV/HA pseudovirions in a dose-dependent manner by specifically interfering with NA activity (see Fig. 1). This work demonstrates the versatility of the assay we have developed to study NA function as a useful platform to evaluate NA sequence variants exhibiting altered sensitivity to oseltamivir carboxylate. This simple system provides a surrogate assay for NA function which may allow us to screen for novel therapeutics against highly pathogenic influenza viruses such as the 1918 Spanish flu and current H5N1 bird flu.
Class Ⅰ: oseltamivir-resistant NAH substitutions. N294S and H274Y substitutions of NA demonstrate the reduced susceptibility to oseltamivir. The oseltamivir-resistant phenotype is demonstrated by high luciferase activity and HA titers that is consistent with the phenotype of parental NA. There is a decrease in luciferase activity and HA activity observed for both H274Y and N294S at 1 μM of oseltamivir. Contrary to the oseltamivir-resistant phenotype, we speculate that this is a result of oseltamivir-mediated inhibition of HIV/HA pseudoviral particle release from 293T producer cells. At 1 μM concentration, the enzyme active sites may be saturated with oseltamivir despite structural studies demonstrating a reduced NAI binding affinity and an increased dissociation rate (Collins P J, et al., 2008). Saturation of the NA enzyme active site with oseltamivir may impede Neu5Ac access to the receptor-binding domain and thus affect NA enzymatic activity.
Oseltamivir carboxylate is a sialic acid analogue that contains a hydrophobic pentyloxy substituent in place of the polar glycerol group at the C6 position. This modification requires a structural rearrangement of the enzyme active site in order to accommodate oseltamivir binding. Structure-based analysis demonstrates that both H274Y and N294S hinder the necessary structural changes to accommodate NAI binding and thus, the weaker interactions leads to inhibitor resistance while stably maintaining the residue substitutions (Collins P J, et al., 2008; Russell R J, et al., 2006).
Class Ⅱ: oseltamivir-sensitive NAH substitutions. The NA substitutions D198N and I222L remained sensitive to oseltamivir treatment and demonstrated a similar dose-dependent inhibition of HIV/HA pseudoviral particle release to NA. In the presence of 1 nM oseltamivir there was no effect on resulting luciferase activity suggesting that HIV/HA pseudovirions were efficiently released from the 293T producer cells regardless of the presence of oseltamivir. The binding affinity of oseltamivir for the NA substitution mutants has yet to be determined; however, we speculate that Neu5Ac may outcompete inhibitor for binding within the receptor-binding pocket at the low nanomolar concentration. A decrease in luciferase activity is observed at 10 nM oseltamivir and the decrease was more pronounced for the D198N substitution compared to I222L. Thus, D198N is more sensitive to oseltamivir and exhibits a greater inhibition of HIV/HA pseudoviral particle release perhaps due to a greater binding affinity.
Mutant E119V displayed properties of both Class Ⅱ and Class Ⅲ mutants. It yielded lower levels of luciferase activity in the absence of oseltamivir indicating a compromised NA enzymatic activity. This suggests that the substitution may destabilize the tetramer, an observation that has been reported for NA variant E119G in NA/N9 (Colacino J M, et al., 1997). Despite its impaired function, we demonstrate that E119V is susceptible to oseltamivir. In the presence of 1 and 10 nM oseltamivir, luciferase levels are similar to that observed for E119V in the absence of inhibitor, however, at 100 nM and 1 μM there is a dosedependent decrease in luciferase activity.
Our findings suggest that E119V is sensitive to oseltamivir treatment. Previous reports have demonstrated a variety of substitutions at position 119 in NA N2 arising from oseltamivir-treatment (Baz M, et al., 2006; Kiso M, et al., 2004; Whitley R J, et al., 2001). Additionally, D198N and I222T substitutions in NA are associated with influenza B virus resistance to oseltamivir (Hatakeyama S, et al., 2007; Ison M G, et al., 2006). Recombinant H3N2 viruses carrying individual D198N and I222L mutations resulted in reduced sensitivity to oseltamivir (Richard M, et al., 2008). A multi-variant NA (H3N2) isolate from an immunocompromised child treated with oseltamivir contained E59G, E119V, and I222V mutations and the I222V mutation was demonstrated to enhance the level of resistance that was originally conferred by the E119V substitution (Baz M, et al., 2006). The results of our study demonstrate that NA substitution of catalytic residues conferring oseltamivir-resistance is subtype-specific.
Class Ⅲ NA substitutions exhibiting a defective phenotype. R292 is conserved among all viral and bacterial NAs and interacts with the carboxylate group of Neu5Ac and NAI substrates positioned within the active site (Crennell S J, et al., 2000; Crennell S J, et al., 1993). Substitution of arginine with a lysine, despite having similar side-chain properties, results in a drastically distinct phenotype which is dependent upon the context of the NA subtype. R292K is associated with NAI-resistance. A/H3N2 clinical isolates recovered from oseltamivir-treated individuals contained NA R292K (McKimm-Breschkin J, et al., 2003; Tai C Y, et al., 1998). NA N2 viruses containing the R292K substitution also display reduced susceptibility to zanamivir (Gubareva L V, et al., 1997). Oseltamivir-resistant viruses may differ substantially in virus fitness and transmissibility (Herlocher M L, et al., 2002; Herlocher M L, et al., 2004; Yen H L, et al., 2005).
In this study we have demonstrated that NA R292K, and to a lesser extent, E119V, were severely compromised in HIV/HA pseudoviral particle release from 293T producer cells. R292K resulted in background luciferase levels indicating that the substitution caused a defect to the NA structure that adversely affected its enzymatic activity. We speculate that the introduction of a lysine, though it may accommodate substrate interactions with the carboxylate group, has a localized effect disrupting stabilizing interactions associated with neighboring residues. The same stabilizing interaction(s) it appears is not necessary for protein stability of NA N2 and instead the R292K mutation is accommodated and supports inhibitor resistance.
Investigation of NA variants with altered drug susceptibility has led to mounting information in the literature. Although N294S is associated with oseltamivir resistance for both NA N1 and N2 subtypes, it does not appear to be the case for all NA substitutions. The residues lining the active site participate in important interactions stabilizing the pocket and may play an essential part in structural changes that are necessary to support NAI binding and catalysis. Thus, highly conserved catalytic residues from different NA subtypes seemingly play different roles. In the case of E119V and R292K mutations, these substitutions are more likely accommodated in NA N2 than in NA N1, as we have demonstrated, whereas resistance associated with D198N and I222L in NA of influenza B viruses demonstrates sensitivity in NA N1. Consistent with previous reports, our findings confirm that H274Y, which has been exclusively isolated in NA N1, displayed a resistant phenotype. Resistance was also demonstrated with N294S, which has been associated with both NA N1 and N2 subtypes.
Particular NA mutations appear to be more prevalent than others, but the frequency at which these specific mutations arise as a result of drug treatment remains uncertain. Recently, mutations V116 and I117 were identified in H5N1 viruses from chickens and were associated with resistance to oseltamivir indicating that new substitutions continue to arise that confer resistance to current NAIs (Hurt A C, et al., 2007). However, when tested in an H1N1 background both were susceptible to oseltamivir. These finding present interesting implications for NA substitutions associated with NAI-resistance not only among subtypes but also the implications of the impact of strain-specific mutations. Resistance-associated NA mutations can cause different levels of functional loss of NA of the same subtype (Yen H L, et al., 2007).
The current study focuses only on the effects of oseltamivir and it would be interesting to also evaluate these NA substitution mutants in the presence of other NAIs including zanamivir and peramivir (Babu Y S, et al., 2000; Maring C J, et al., 2005; Stoll V, et al., 2003). A promising new NAI in the early stages of drug development is A-315675—a pyrrolidine-based compound developed from Abbott Laboratories— that has successfully demonstrated effective in vitro activity against influenza A (N1, N2, and N9 subtypes) and B viruses with similar efficacy to that of zanamivir and oseltamivir (DeGoey D A, et al., 2002; Kati W M, et al., 2002). However, N1 NAs containing H274Y and N294S mutations have been demonstrated resistance to A-315675 (Abed Y, et al., 2008). Thus, it seems practical to expand efforts to develop anti-influenza inhibitors that target new NA protein regions. The expansive repertoire of NA structure information, which includes nearly all NA subtypes, provides new opportunities for drug design (Russell R J, et al., 2006; Taylor G, et al., 1993).
In summary, this study demonstrates that the HIV-based pseudotyping system provides a quick, safe, and effective way to assess resistance to neuraminidase inhibitors. And we believe that as new mutations appear in influenza isolates, their impact on the effectiveness of current and future anti-NA can be quickly and reliably evaluated by this assay.














 DownLoad:
DownLoad: